Single cell profiling reveals window for immunotherapy in liver cancers
Primary liver cancer has the second highest mortality rate with more than 700,000 deaths. Hepatocellular carcinoma (HCC), the most prevalent tumor type in the liver, arises predominantly from hepatocytes and is often related to known etiologies such as chronic hepatitis B or C virus, obesity, diabetes mellitus, metabolic liver diseases and life style factors (for example, alcohol). The injury to the liver parenchyma, these risk factors induce, causes an inflammatory milieu, which initiates a chain of accumulative events by producing reactive oxygen species (ROS), inflammatory cytokine and chemokine release as well as chromosomal instability. In this milieu, progressions of cellular death (necrosis) and regeneration result in liver fibrosis. In the background of the damaged liver, molecular alterations accrue contributing to the initiation of HCC. This multi-step progression and a detailed understanding of the genetic and epigenetic events that define HCC development remain challenging. The severity of this malignancy is further complicated since it commonly is associated with underlying cirrhosis, which clinically is problematic for the advancement of therapeutic options.
Recent studies have emphasized the molecular complexity in primary HCC, characterizing distinct interpatient tumor heterogeneity, including prevalent mutations in genes, such as TP53, CTNNB1, chromatin remodeling factors, and frequent early variants in the TERT promoter (1,2). Regardless of these targets, patients often response different to treatment or markedly show drug resistance. As such, essentially the last decade of clinical phase III trials has failed and the only approved therapy is the multi-tyrosine kinase inhibitors (sorafenib and regorafenib). This genomic tumor complexity is further underlined by a prominent intra-tumor heterogeneity (ITH) (3,4), which is defined as the morphologic diversity, differential phenotypic potential, and genome variance of distinct tumor cells comprising a single tumor mass. The model of “branched evolution” has emerged as a reflection of this tumor heterogeneity, including both the molecular and cellular convolution of the disease (tumor mass) that complicates accurate patient characterization and stratification as well as clinical management. HCC is typically innately resistant to treatment at diagnosis and from a therapeutic perspective, the heterogeneous branched evolution suggests only a partial sampling of a given tumor mass with apparent consequences to precision-based clinical decisions, and thus, limited or incomplete drug response.
In the liver, this scenario is broadened by a complexity in the putative “cell of origin” leading to malignant transformation, including evidence of cellular dedifferentiation resulting in the hepatocytic lineage to give rise to tumor formation in the intrahepatic bile ducts (5,6), suggesting a marked continuum both genotypic and phenotypic of the malignant cells in the liver (7). This quandary is further complicated by an anticipated outgrowth of dormant cancer cells that present a distinctly different drug response or adaptation to therapy.
Recent integromics studies, including The Cancer Genome Atlas (TCGA) (8) and Thailand Initiative in Genomics and Expression Research for Liver Cancers (TIGER-LC) (9) have both characterized new patient subclasses and drug targets. Moreover, these consortia-led approaches have additionally stressed the potential for introducing immunotherapy into liver cancer management. As such, 22% (43/196) of the HCC cases presented by histopathological evaluation (TCGA core patient set) show high to moderate levels of tumor infiltrating lymphocytes (TIL). Hierarchical clustering of patient-based gene expression of immune surface markers outlined two groups with elevated expression of key immune checkpoint genes CTLA-4, PDCD1 and CD274/PD-L1. Irrespective of viral disease status (hepatitis B or C viral infection), a significant difference in the immune cell composition was determined between tumor and the adjacent surrounding liver, with the tumor microenvironment (TME) demonstrating a change in the immune cell composition from activating effector cells to resting suppressive immune cells. This includes depletion of naïve B cells, neutrophils, monocytes and gamma/delta T cells as well as enrichment of memory B cells, regulatory T cells (Tregs), resting dendritic cells (DCs) and undifferentiated (M0) macrophages.
Given the importance of TME for the main clinical hurdles (immunosuppression, chemoresistance and metastasis) in both localized and non-localized disease, targeting the immune compartment of the tumor mass has noticeably altered clinical oncology. Predictive factors of response to immunotherapy include mutational burden, TIL levels, and expression of the markers CTLA-4, PDCD1 and PD-L1. Information about the tumor-residing T cells is dependent on and, as such, vital for understanding the efficacy of immunotherapy. The premise of immune treatment is in the TME to release a cytotoxic response, which is elicited by CD8+ T cells, an activity that often is abrogated either through suppression of Tregs or T cell exhaustion (abrogated T cell effector function). In this view, though HCC often has high levels of TILs, this tumor type remains immunologically challenging since the TIL function/effect is deregulated.
In work recently presented in Cell (10), Zheng and colleagues provided a comprehensive transcriptomic and T cell receptor (TCR) map of more than 5,000 single tumor-infiltrating T cells isolated by FACS from blood, adjacent normal and tumor tissues obtained from six individual HCC patients. Remarkably, a total of 11 unique T cell subsets were elucidated by transcriptomics, defining 5 CD8+ (cluster C1–C5) and 6 CD4+ (cluster C6–C11) sub-clusters represented by distinct gene signatures. Importantly, each of these T cell subsets revealed a tissue-specific distribution with additional evidence of developmental gene expression connectivity. Notably, utilizing SC3 clustering each of the major clusters were dominated by a unique T cell representation. The functional presentation emphasized characteristics of each cluster with C1 expressing naïve marker genes (for example, LEF1) associated with peripheral blood, C2 distinguished by T cell effector function, C3 comprising genes (SLC4A10) related to mucosal-associated invariant T (MAIT) cells prevalent in the adjacent normal liver tissue whereas C4–C5 predominantly are composed of tumor-associated CD8+ exhausted T cells represented by markers CTLA4, PDCD1 and differential regulation of the GZMK expression signature. The residual six clusters (C6–C11) were dominated by CD4+ T cells with C6 primarily originating from peripheral blood, C7–C8 both represented FOXP3+ Tregs (CD4+/CD25high) with either of the sub-clusters main expression pattern involved in blood or tumor tissue, respectively. CD4+ Tregs are further defined by key expression of genes such as FOXP3, TNFRSF9, TIGIT and CTLA4. The last three clusters (C9–C11) are predominantly associated with T helper cell gene expression characteristics with C10 associated to exhausted CD4+ cells (CXCL13, PDCD1, CTLA4, TIGIT) and elevated cytotoxic marker-gene expression (NKG7, GNLY, GZMB) in cluster C11. This study presents for the first time a comprehensive evidence for a differential T cell distribution in the TME of HCC, revealing both known and novel marker genes associated with TILs specifically underlining the presence of exhausted CD8+ T cells and Tregs by for example marker genes CCR8 and LAYN. Interestingly, the gene LAYN is associated with exhausted CD8+ TILs and poor prognosis. Upon T cell (CD8+ and Treg) activation LAYN is elevated, whereas LAYN expression attenuates the expression of interferon gamma by CD8+ T cells, which is required for active tumor cell killing (Figure 1).
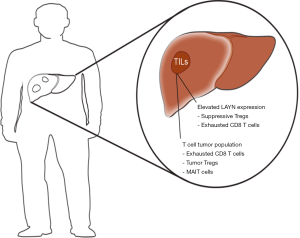
In HCC, the TME presents an augmented existence of exhausted CD8+ T cells and Tregs. Importantly, clonal TCR findings suggest that tumor-associated Tregs are recruited since 82% of Tregs in the tumor are unique, and thus, do not show evidence of evolution directly from the tumor-residing CD4+ T cells or expansion of Treg derived from the tumor-adjacent normal tissue. Contrary, exhausted CD8+ T cell clones demonstrated similarity with 37% shared between CD8+ clusters (C1–C5), which suggests a transformation of T cells within the tumor into exhausted CD8+ T cells. As such, C5 (tumor-associated exhausted CD8+ T cell expression) was proposed to present a possible cluster/profile harboring T cell expression signaling that may be suggestive of a transition from effector to exhausted T cells within the tumor. These cells may be candidates for therapeutic targeting, stimulating their effector T cell-like phenotype to revive their T cell function and avoid regulatory exhaustion. Additional, C9 was highlighted as a putative route for immunotherapy to promote cytolytic CD4+ T cell function. Current clinical trials, which utilize immunotherapy in HCC, include Keynote-224 (phase II to estimate activity and toxicity of pembrolizumab, n=100), another phase II trial will investigate pembrolizumab in advanced unresectable HCC cases, CheckMate-040 (phase I–II to determine the safety of nivolumab) and CheckMate-459 (phase III trial that will investigate nivolumab vs. sorafenib with primary endpoints overall survival and time to progression, n=726) (11).
In conclusion, the purpose of immunotherapies is essentially to modify the TME both within the primary tumor site and metastatic niche(s) in order to increase the level of active tumor-specific CD8+ effector cells relative to suppressive T cells, inflammatory cells and their mediators. Additional, the goal is to induce a systemic memory eliciting an anti-tumor immune-response and thus, to control residual tumor cells. In the current study, the authors have defined a comprehensive map of the circulating and tumor-residing T cells, underscoring their unique genomic characteristics and emphasizing a path for immunotherapy in HCC.
Acknowledgements
Funding: JB Andersen is supported by grants from the Novo Nordisk Foundation, Danish Cancer Society, Danish Medical Research Council and European Commission for Research and Innovation (No. 14040; R98-A6446; 4183-00118A).
Footnote
Conflicts of Interest: The author has no conflicts of interest to declare.
References
- Nault JC, Mallet M, Pilati C, et al. High frequency of telomerase reverse-transcriptase promoter somatic mutations in hepatocellular carcinoma and preneoplastic lesions. Nat Commun 2013;4:2218. [Crossref] [PubMed]
- Schulze K, Imbeaud S, Letouze E, et al. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat Genet 2015;47:505-11. [Crossref] [PubMed]
- Birkbak NJ, Andersen JB. Heterogeneity Among Liver Cancer--A Hurdle to Optimizing Therapy. Gastroenterology 2016;150:818-21. [Crossref] [PubMed]
- Xue R, Li R, Guo H, et al. Variable Intra-Tumor Genomic Heterogeneity of Multiple Lesions in Patients With Hepatocellular Carcinoma. Gastroenterology 2016;150:998-1008. [Crossref] [PubMed]
- Holczbauer A, Factor VM, Andersen JB, et al. Modeling pathogenesis of primary liver cancer in lineage-specific mouse cell types. Gastroenterology 2013;145:221-31. [Crossref] [PubMed]
- Zender S, Nickeleit I, Wuestefeld T, et al. A critical role for notch signaling in the formation of cholangiocellular carcinomas. Cancer Cell 2013;23:784-95. [Crossref] [PubMed]
- Marquardt JU, Andersen JB, Thorgeirsson SS. Functional and genetic deconstruction of the cellular origin in liver cancer. Nat Rev Cancer 2015;15:653-67. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive and Integrative Genomic Characterization of Hepatocellular Carcinoma. Cell 2017;169:1327-41.e23. [Crossref] [PubMed]
- Chaisaingmongkol J, Budhu A, Dang H, et al. Common Molecular Subtypes Among Asian Hepatocellular Carcinoma and Cholangiocarcinoma. Cancer Cell 2017;32:57-70.e53. [Crossref] [PubMed]
- Zheng C, Zheng L, Yoo JK, et al. Landscape of Infiltrating T Cells in Liver Cancer Revealed by Single-Cell Sequencing. Cell 2017;169:1342-56.e16. [Crossref] [PubMed]
- Cidon EU. Systemic treatment of hepatocellular carcinoma: Past, present and future. World J Hepatol 2017;9:797-807. [Crossref] [PubMed]


