Window-of-opportunity: neonatal gut microbiota and atopy
The microorganisms within the gastrointestinal tract have received considerable attention in recent years as new tools and faster, less expensive genomics approaches, have allowed for associations between microbial behavior and disease (1). Colonization of our GI tract occurs at birth and remains unstable until approximately 3 years of age, a stage of life during which time major physiological systems such as the immune system also develop (2). Longitudinal tracking studies have suggested that this early period is critical in predisposing us to conditions, such as asthma (2,3). By profiling the microbial community using 16s rRNA gene sequencing, Fujimura et al. showed that neonates could be grouped into three broad microbiota conformations. One of these conformations was significantly associated with higher risk of developing atopy, an allergic hypersensitization predisposing individuals to asthma (4) but was only detectable in children under 6 months of age. Furthermore, by using sterile fecal water in T cell polarization assays, Fujimura et al. linked the microbial state with a functional readout relevant to disease (4). This is significant because it identifies a potential window-of-opportunity to identify at-risk individuals and, if a causative mechanism can be revealed, intervene prophylactically to reduce susceptibility.
Fujimura et al. studied a cohort of 298 children whose fecal bacterial and fungal communities were analyzed at one month of age (n=130, termed “neonates”) and/or at six months of age (n=168, termed “infants”). Consistent with previous studies, the diversity of the bacterial community within each infant (alpha-diversity) increased with age, while the alpha-diversity of the fungal community decreased (5). An inverse correlation which may suggest that inter-kingdom competition is relevant to colonization. Speaking in broad terms, this represents a transition from a state dominated by Bifidobacteriaceae and Enterobacteriaceae families in the neonate towards one including larger proportions of Bacteroidiaceae and Lachnospiraceae in the infant, and a contraction of Malasseziales within the neonatal fungal community leaving the infant community dominated by Saccharomycetales (Figure 1). Between individual children, the bacterial and fungal communities became increasingly different to each other with age, indicative of the establishment of a diverse community which is, to large degree, specific to individuals.
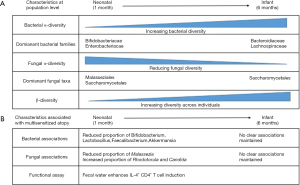
To identify signatures associated with atopy, Fujimura et al. first applied a modelling program to determine if any of the neonatal diversity could be explained by separating the population into sub-groups. They found that three sub-groups (NGM1, NGM2 and NGM3) offered the best distinction. NGM3 (n=11 samples) showed depletion of Bifidobacteria, Lactobacillus, Faecalibacterium and Akkermansia, and an increased relative risk of multisensitised atopy at age 2 years and parental-reported doctor-diagnosed asthma at age 4 years. Alterations in the fungal community were also characteristic of the NGM3 state, with a reduced proportion of Malassezia, and increased proportions of Candida and Rhodotorula. A separate study analyzing the fecal microbiota of children aged 3 months found similar depletion of bacterial taxa to be associated with atopy (6), and bacterial depletion by early-life antibiotic treatment has been associated with allergic asthma development in mice and humans (2,3) increasing confidence in these associations. Furthermore, loss of these taxa has been observed in other immune-mediated conditions, raising the question as to whether there are beneficial features about our relationship with these microbes that translate across diseases (7,8). At 6 months of age the infant microbiome no longer fitted to these three states, but rather fitted two that did not associate with multi sensitized atopy. This leads the authors to suggest that analysis in the neonatal period is critical to identify microbiome associations, a hypothesis consistent with previous data (2,6).
Microbiome research is often criticized for over-reliance on-omics based associations rather than biological function. Fujimura et al. correlated the NGM3 state with changes in the metabolite signature of fecal water, and assessed the functional impact of this signature on T cell polarization in vitro. Consistent with these metabolites having immunomodulatory function predisposing to allergy, the fecal water from children classified as NGM3 slightly, but significantly, enhanced the induction of IL-4+ CD4+ T-cells, compared to fecal water from children classified as NGM1. Fecal water from both NGM1 and NGM3 states inhibited the induction of anti-inflammatory FOXP3+ CD4+ T cells, which is not surprising as both would contain pro-inflammatory microbial ligands, such as LPS. Of interest was the finding that the metabolite, 12, 13-DiHOME, was enriched in the NGM3 state, and treatment of dendritic cells in vitro resulted in a suppression of FoxP3+ T cell induction.
While the data presented in this study are promising, many questions remain. Although the relative risk for multi-sensitized atopy was higher in neonates identified as NGM3, infants identified as NGM1 (13 out of 70) and NGM2 (13 out of 49) still developed the condition, and a significant proportion of those identified as NGM3 did not develop disease (5/11). Comparing atopy based on non-specific IgE levels (>0.35 IU/mL) also showed no association with NGM state. Thus, it is not true that having the microbiome association described here is necessary or sufficient for disease. Given the instability of the microbiome in early life, it’s possible that a single time-point may not reflect significant periods in which the microbiome may fluctuate in different states. Our immune response likely reflects the net outcome of our interactions with the microbiome during this period, thus better understanding early life dynamics is critical (2). It is also possible that there is functional redundancy between species, so that community states appearing distinct on a taxonomic level, display more similarity when analyzed on a functional level. It will be interesting to determine if combining taxonomic data with functional assays, such as the cellular response to fecal water, increases the predictive power of analyzing the microbiome for multi-sensitized atopy.
In conclusion, the work by Fujimura et al. highlights that analyzing the composition of the microbiome in the neonatal period could be critical to identifying associations with multi sensitized atopy. Although much work needs to be done for this to be useful therapeutically, it offers important first insight and a framework for future study.
Acknowledgements
None.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Blumberg R, Powrie F. Microbiota, disease, and back to health: a metastable journey. Sci Transl Med 2012;4:137rv7. [Crossref] [PubMed]
- Nylund L, Satokari R, Salminen S, et al. Intestinal microbiota during early life - impact on health and disease. Proc Nutr Soc 2014;73:457-69. [Crossref] [PubMed]
- Stiemsma LT, Turvey SE. Asthma and the microbiome: defining the critical window in early life. Allergy Asthma Clin Immunol 2017;13:3. [Crossref] [PubMed]
- Fujimura KE, Sitarik AR, Havstad S, et al. Neonatal gut microbiota associates with childhood multisensitized atopy and T cell differentiation. Nat Med 2016;22:1187-91. [Crossref] [PubMed]
- Yatsunenko T, Rey FE, Manary MJ, et al. Human gut microbiome viewed across age and geography. Nature 2012;486:222-7. [PubMed]
- Arrieta MC, Stiemsma LT, Dimitriu PA, et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci Transl Med 2015;7:307ra152. [Crossref] [PubMed]
- Miquel S, Martín R, Rossi O, et al. Faecalibacterium prausnitzii and human intestinal health. Curr Opin Microbiol 2013;16:255-61. [Crossref] [PubMed]
- Tojo R, Suárez A, Clemente MG, et al. Intestinal microbiota in health and disease: role of bifidobacteria in gut homeostasis. World J Gastroenterol 2014;20:15163-76. [Crossref] [PubMed]


