
Long noncoding RNA HOTTIP expression predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation
Introduction
Hepatocellular carcinoma (HCC) is one of the most common malignant neoplasms worldwide and causes approximately 700,000 deaths each year (1). Half of these cases and deaths were reported to occur in China and were attributed to the high prevalence of hepatitis B virus infections and liver cirrhosis (2,3). As a potentially curative treatment for liver cirrhosis and cancer, liver transplantation (LT) is widely used to treat selected patients with early-stage HCC, and it exhibits the greatest benefit among all selectable therapeutics (4,5). However, the frequent recurrence and high metastasis rates of HCC hinder positive outcomes for patients. Because multiple genetic alterations are responsible for the progression of HCC (6), exploration of the molecular mechanism of HCC is important for disease diagnosis, treatment, and prediction of outcomes.
Recently, long noncoding RNAs (lncRNAs), noncoding RNAs that range from 200 nucleotides up to approximately 100 kb in length, have been presumed to play a significant role in various cellular processes, including immune surveillance, cell-cycle regulation, stem cell pluripotency and tumorigenesis (7,8). The lncRNA HOTTIP, which is involved in the Hox gene network, could affect cell function by targeting the transcription of HOXA genes (9), which can cause epigenetic alterations and promote tumorigenesis and tumor progression (10,11). Recent studies have described HOTTIP as a prognostic marker that plays a key role in promoting tumorigenesis and cancer development in HCC (12-14) and pancreatic cancer (15) .
The Milan criteria for identifying candidate HCC patients for LT have been accepted worldwide. However, they ignore the molecular biomarker expression patterns in HCC and have been criticized for being too restrictive. Research groups at our center continually aim to explore more inclusive criteria for identifying LT candidates in the HCC patient population, which might help more advanced HCC patients have the opportunity to receive LT treatment and obtain a similar prognosis compared to the Milan criteria (16,17). In the present study, we investigated the expression pattern and single nucleotide polymorphism (SNP) genotype of HOTTIP in HCC tissues from patients who received transplant therapy and its relationship to clinicopathological parameters to explore the utility of HOTTIP in predicting tumor recurrence in HCC patients after LT. We also explored the functional implications of HOTTIP in HCC using in vitro assays.
Methods
Ethical statement
This study was approved by the Medical Ethics Committee of the First Affiliated Hospital of Zhejiang University (NO. 2016209), and informed consent was obtained from all patients.
Patient samples
A total of 155 HCC patients treated with LT from 2003 to 2010 at our hospital (First Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou, China) were enrolled in the present study. The inclusion criteria of patients were as previously described of our other study (18). Especially, all patients were HBV-positive (HBsAg+) and none of them was hepatitis C virus (HCV)-positive. This study was approved by the Medical Ethics Committee of the First Affiliated Hospital of Zhejiang University, and informed consent was obtained from all patients. These patients were diagnosed with HCC either before or after transplantation. The diagnosis was confirmed by histopathological examination, and complete clinical and laboratory data were available prior to surgery and during follow-up. The follow-up course and diagnostic criteria of recurrence have been described previously (19). The distribution of clinicopathologic data in the study cohort is given in Table 1. Specimens of cancer/noncancerous tissues and clinical information were available from these patients after obtaining informed consent.
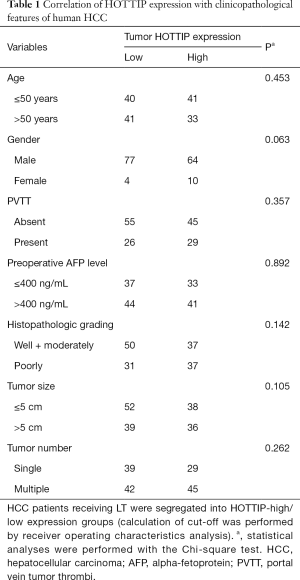
Full table
Postoperative follow-up
The surviving patients were followed up at the outpatient clinic. The median follow-up was 25.45 months. Tumor recurrence was monitored by alpha-fetoprotein (AFP), ultrasonography, chest X-ray, and Emission computed tomography every 3 months for the first 2 years and semiannually thereafter. Imaging techniques, either intrahepatically or extrahepatically (lymph nodes, distant metastases) were used diagnosing recurrence, however, a simple increase of AFP was not diagnosed as recurrence.
Genotyping
Genomic DNA was isolated from peripheral blood samples of 102 HCC patients (involved in the 155 patients underwent LT) DNeasy Blood and Tissue Kit (Qiagen, Valencia, CA) according to the Manufacturer’s instructions. Briefly, cell pellets from 1 mL peripheral blood samples were plated in microtubes and supplemented with 200 µL working solution (20 µL proteinase K stock solution and 180 µL Buffer ATL). After repeated mixed, the mixture was incubated at 56 °C until completely lysed. Adding 410 µL Buffer AL–ethanol mixture in the microtubes and shaking the racks vigorously up and down for 15 s, then the solution was collected and transferred to DNeasy 96 plates. After centrifuging for 10 min at 6,000 rpm, the samples were respectively dealt with 500 µL Buffer AW1 and AW2, and then supplemented with 200 µL Buffer AE. Finally, the DNA was stored for the next experiment after centrifuge for 2 min at 3,800 g. And the SNaPshot was employed to analyze the type of SNPs in HOTTIP.
Cell lines and chemical reagents
The normal liver cell line LO2; the liver cancer cell lines SMMC-7721, Huh-7, HepG2, Bel-7402, SK-Hep1, PLC/PRF/5, and Hep3B; and the metastatic human HCC cell lines HCCLM3, MHCC97L and MHCC97H were purchased from the American Type Culture Collection (Manassas, VA, USA), the Shanghai Institute of Cell Biology (Shanghai, China), and the Liver Cancer Institute of Fudan University (Shanghai, China). All cell lines were maintained under the recommended culture conditions and incubated at 37 °C in a humidified environment containing 5% CO2. The chemical reagents, including doxorubicin, etoposide and oxaliplatin, were obtained from Sigma-Aldrich (St. Louis, MO, USA).
Real-time quantitative reverse-transcription polymerase chain reaction (qPCR)
Total RNA was extracted from cell lines or cancerous/noncancerous specimens by using trizol (Invitrogen, CA, USA) and miScript II RT Kits (Qiagen, Valencia, CA), and cDNA was synthesized. The expression levels of HOTTIP and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were determined by qPCR using the following primer sequences:
The qPCR reactions were performed with the ABI7500 system (Applied Biosystems, CA, USA) and SYBR Green dye (TaKaRa Biotechnology, Dalian, China). GAPDH was used as an internal control. The HOTTIP values were normalized to GAPDH. ΔΔCT analysis was used to analyze the PCR data.
RNA interference
SMMC-7721, BEL-7402 and Huh-7 cells were transfected with 50 nM siRNAs targeting HOTTIP or siGFP (GenePharma, Shanghai, China) using Lipofectamine 2000 transfection reagent (Invitrogen, CA, USA) according to the manufacturer’s instructions. The siRNAs sequences were:
After 48 h, the knockdown of HOTTIP was confirmed by real-time PCR.
Analysis of cell viability and invasiveness
After 48 h of RNA interference and 24 h of exposure to various agents, viable cells that were not stained with trypan blue (0.4%) were counted to measure cell viability. A cell-invasion assay was performed using a Transwell (Millipore, Billerica, MA, USA)-based method. Forty-eight hours after RNA interference, 5×104 cells were resuspended in FBS-starved media and seeded on the filters coated with Matrigel (BD Bioscience) in the upper compartment, and the lower compartment was filled with media containing 15% FBS. After 24 h, the cells on the bottom surface were fixed with methanol and stained with 0.1% crystal violet. The cells were counted and photographed using digital microscopy. Three fields were counted per filter in each group. The experiments were performed in triplicate.
Human gene expression microarrays (Affymetrix U133 Plus 2 chip)
Human gene expression microarrays (Affymetrix U133 Plus 2 chip) were used to analyze the differential gene expression in response to 48 h of siHOTTIP or siGFP treatment in SMMC-7721 cells. Cluster analysis was performed based on Gene Ontology and KEGG to find differentially expressed gene families or pathways.
Statistical analysis
Continuous data were analyzed by the Student t-test to assess differences between the 2 groups. Categorical data were analyzed by the Chi-square test. The recurrence-free survival probability was analyzed with the Kaplan-Meier method, and a log-rank test was used to estimate the differences between groups. Independent prognostic indicators were determined in the multivariate analysis using Cox’s proportional hazard model. The expression correlation was analyzed by linear spearman rank test (2-tailed). All statistical analyses were performed using SPSS (Statistical Package for the Social Sciences) 16.0 (SPSS, Chicago, IL, USA) and GraphPad Prism 5.0 (GraphPad Software, La Jolla, CA, USA). P<0.05 was considered to indicate significant differences.
Results
Expression of HOTTIP in liver cancer cell lines and the tissues of HCC patients
We measured the expression levels of HOTTIP in a human normal liver cell line LO2 and ten liver cancer cell lines by quantitative real-time RT–PCR. Of the 10 liver cancer cell lines, 4 (Huh-7, MHCC-97L, MHCC-97H, and BEL-7402) expressed higher levels of HOTTIP than the normal liver cell line (Figure 1A). Additionally, the relative expression levels of HOTTIP RNA in tumors and adjacent non-tumorous tissues from 155 HCC patients were also measured. Tumors expressed significantly higher levels of HOTTIP than adjacent non-tumorous tissues. Furthermore, recurrent tumors expressed more HOTTIP than non-recurrent tumors (P<0.01, Figure 1B).
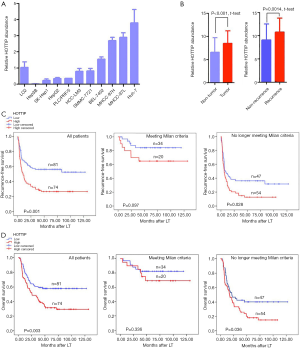
High HOTTIP expression is associated with a poorer prognosis in HCC patients
To explore the utility of HOTTIP as a promising molecular marker to predict the prognosis of HCC patients, we compared the recurrence-free survival times and overall survival times between 155 HCC patients who expressed high or low expression levels of HOTTIP RNA based on extensive clinical follow-up data. First, HOTTIP expression did not correlate with any single clinicopathological characteristic in the 155 patients, including age, gender, PVTT (portal vein tumor thrombi), AFP (alpha-fetoprotein), histopathologic grade, tumor size or tumor number, when stratified by expression level (high or low) (Table 1). Furthermore, the univariate recurrence-free survival analysis showed that the tumor size, preoperative AFP level, PPTV, Milan criteria and HOTTIP expression act as risk factors for tumor recurrence after LT (Table 2). In addition, COX multivariate analysis showed that in patients who beyond the Milan criteria, the PPTV, a tumor size >5 cm and a high expression of HOTTIP are the independent risk factors for tumor recurrence after LT and predict shorter overall survival times (Table 3).
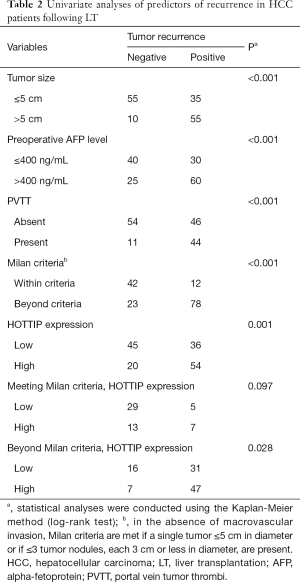
Full table
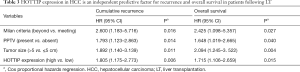
Full table
The overexpression of HOTTIP predicted earlier recurrence and shorter overall survival times in HCC patients who underwent LT (P=0.001, Figure 1C, P=0.003, Figure 1D). Our results revealed that the Milan criteria effectively predicted the risk of tumor recurrence (Table 3, Beyond vs. Meeting, HR =2.600, P=0.016) and overall survival times (Table 3, Beyond vs. Meeting, HR =2.425, P=0.027) in patients after LT, which is consistent with previous studies (20). Interestingly, in patients who met the Milan criteria, further classification based on HOTTIP expression did not result in significant differences in the recurrence-free survival times (P=0.097, Figure 1C) or overall survival times (P=0.336, Figure 1D). However, patients who beyond the Milan criteria and who exhibited increased HOTTIP expression levels experienced earlier tumor recurrence and lower overall survival rates following LT (P=0.028, Figure 1C, P=0.036, Figure 1D). Taken together, our clinical findings indicate that HOTTIP expression is an important indicator for predicting tumor recurrence and overall survival times in HCC patients after LT, which may also be valid in patients who beyond the Milan criteria.
The effects of HOTTIP RNA expression on malignant biological behaviors of hepatocellular carcinoma cells
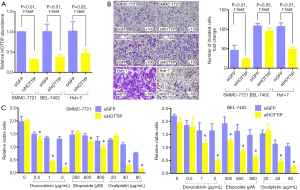
SMMC-7721, BEL-7402 and Huh-7 cells, the hepatocellular carcinoma cell lines that express high levels of HOTTIP, were transfected with siRNAs targeting HOTTIP. Consequently, HOTTIP expression was down-regulated (Figure 2A). An in vitro Transwell assay showed that the down-regulation of HOTTIP expression could significantly reduce the invasiveness of hepatocellular carcinoma cells (Figure 2B). The down-regulation of HOTTIP in HCC cells significantly enhanced the sensitivity of tumor cells to Doxorubicin, Etoposide and Oxaliplatin (Figure 2C).
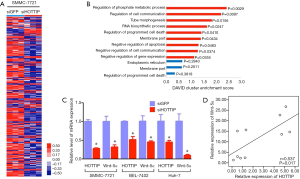
The impact of HOTTIP on the expression of downstream molecules
HOTTIP has been shown to be involved in the regulation of cell migration and the invasion of HCC and other malignant biological behavior, but the exact molecular mechanism remained to be elucidated. We knocked down HOTTIP expression in SMMC-7721 cells with siRNA (compared to the negative control, the HOTTIP expression level decreased by 30.3%), and examined the consequent gene expression changes using human gene expression microarrays (Affymetrix U133 Plus 2 chip). The results showed that 235 genes (176 up-regulated and 59 down-regulated genes, P<0.05, representing a 2-fold difference in expression levels, Figure 3A,B) are closely associated with malignant biological behaviors, such as tumor proliferation, apoptosis and invasion. We discovered that the Wnt pathway is an important pathway that is mediated by HOTTIP by analyzing the information provided by Gene Ontology, KEGG pathway and other biological information web sites. Furthermore, the down-regulation of HOTTIP significantly decreased the expression of Wnt-5a in the Wnt pathway in hepatocellular carcinoma cells (Figure 3C). The positive correlation of HOTTIP and Wnt-5α mRNA was indicated by analysis of 10 tissue samples from 10 of the 155 HCC patients underwent LT (Figure 3D). The above results suggest that HOTTIP might affect the malignant biological behavior of HCC cell migration and invasion by regulating the Wnt pathway.
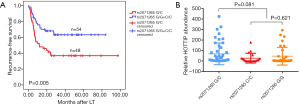
HOTTIP rs2071265(G/C) is associated with an earlier recurrence in HCC patients
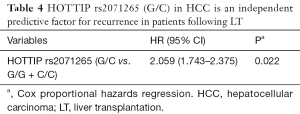
Two SNPs, rs2071265 and rs142077238 in the HOTTIP gene were genotyped in 102 peripheral blood samples. The SNP rs2071265 (G/C vs. G/G + C/C) is associated with an earlier recurrence in HCC patients who underwent LT (P=0.005, Figure 4A), while the SNP rs142077238 of was not associated with HOTTIP expression or patients’ HCC recurrence (data not shown). The SNP rs2071265 had a trend to be associated with HOTTIP expression (P=0.081, Figure 4B). In COX regression analysis, SNP rs2071265 (G/C vs. G/G + C/C) was found to be an independent predictor of HCC recurrence (Table 4, HR =2.059, P=0.022).
Full table
Discussion
LT is a potentially curative treatment for HCC. Despite the development of diagnosis and treatment protocols for HCC in recent decades, the prognosis of HCC remains very poor. HCC frequently recurs after surgery, which is principally attributable to the microscopic extrahepatic metastatic foci before LT, and this recurrence remains the major obstacle for improving the long-term survival rates of HCC patients (21).
HCC is a complex tumor associated with multiple genetic alterations that are responsible for many cancer properties including metastasis (22). A deep exploration of the underlying molecular mechanisms that drive cancer metastasis is highly desirable. The lncRNA HOTTIP was recently identified as a regulator of HOXA genes (9), which could regulate tumor biological behavior (11). A recent study of a cohort of 52 HCC patients reported that the levels of HOTTIP are associated with the clinical progression of HCC patients and predict disease outcomes (12). However, the patient number in the cohort might limit the strength of the conclusion that HOTTIP levels are clinically relevant to the prognosis of HCC patients. Our study of HCC tissues from 155 patients after LT similarly suggested that HOTTIP is a potentially predictive biomarker of HCC. Moreover, patients whose tumors expressed a high level of HOTTIP were at an increasing risk of recurrence post-LT and experience lower overall survival. Multivariate analyses revealed that HOTTIP expression was an independent prognostic factor for tumor recurrence and overall survival rates in HCC patients after LT.
Clinicopathological characteristics, including the tumor grade, tumor size, and vascular invasion, are important predictors of HCC recurrence after LT but remain unsatisfactory for individualized diagnosis and therapy. The Milan criteria, which ignore molecular biomarker expression patterns, have been criticized for being too restrictive, because patients with tumors that beyond the Milan criteria may also have good prognoses (16,20,23). We consider the lack of biological molecular markers to be the major limitation of the Milan criteria. Biological molecules that reflect tumor characteristics or immunity status need to be incorporated into the LT criteria for HCC patients, and our center has continually attempted to address this issue (16-18,24). Certain molecular-level tumor diagnostic methods or prognostic predictions are essential for individualized diagnosis and therapy (25,26). In addition, HCC patients in China, most of whom are infected with the HBV (hepatitis B virus), represent the largest subpopulation of HCC patients in the world (1,27), and the biological characteristics of HBV-induced HCC differ from those of HCV or alcoholic cirrhosis-induced HCC, which frequently occur in western countries (6). Thus, we are dedicated to developing more accurate criteria for LT treatment in HCC patients, which might help more advanced HCC patients have the opportunity to receive LT treatment and a prognosis similar to that of patients who meet the Milan criteria.
Of our study cohort, 20.8% of patients who did not meet the Milan criteria remain tumor-free 5 years after LT, implying that the current Milan criteria could not stratify all patients who were eligible to receive LT. Our results also revealed that patients who beyond the Milan criteria and expressed decreased levels of HOTTIP experienced longer recurrence-free survival times and longer overall survival times, indicating that this patient population may be eligible for LT. This inclusion may significantly increase the number of HCC patients who are candidates for LT. However, a single molecular marker seems insufficient to diagnose HCC or efficiently predict the prognosis (22,28), which is a weakness of this study. The development of high throughout detection techniques has facilitated multi-molecular-level diagnostic or prognostic modeling, which will be a subject of future studies. We also aim to examine more molecular markers and patient samples to establish a prognostic model of recurrence and death in HCC patients after LT and develop a more accurate prognostic criterion for LT treatment in HCC patients. Combined with our previous study (18,24), the present study provides valuable support for the establishment of the model. Our center will begin to preoperatively examine the HOTTIP expression level using liver biopsy specimens or circulating tumor cells (CTC). More institutions will participate in the further study and more patients samples will be involved.
We further detected the genetic variations of HOTTIP in predicting the tumor recurrence of HCC patients underwent LT. The SNP rs2071265 is associated with an earlier recurrence in HCC patients. As far as we know, this is the first investigation of HOTTIP gene polymorphisms on the prognosis of HCC. Interestingly, rs2071265 has no effect on the expression level of HOTTIP Mrna (G/C vs. G/G + C/C, P=0.081). Less of samples involved might be responsible for the negative statistical results. The mechanisms by which SNP rs2071265 affects the function of HOTTIP need further investigations.
Additionally, we explored the biological role of HOTTIP in HCC cell invasion and chemosensitivity using in vitro assays. Knocking down HOTTIP significantly reduced the invasiveness of the HCC cell lines SMMC-7721, BEL-7402 and Huh-7. The down-regulation of HOTTIP also sensitized HCC cells to chemotherapy drugs, such as doxorubicin, etoposide and oxaliplatin. Moreover, whole-genome chip Affymetrix U133 plus 2.0 arrays were used to screen the ability of HOTTIP to regulate downstream genes or pathways. The arrays indicated that HOTTIP may significantly affect the Wnt pathway, which may be involved in the HOTTIP-induced migration and invasiveness of HCC cells. However, while our in vitro assay results imply that HOTTIP plays a malignant biological role, the precise mechanisms by which HOTTIP increases HCC invasiveness or decreases chemosensitivity remain unclear. The relationship between the Wnt pathway and HOTTIP warrants further exploration. Furthermore, the functional changes in HCC cells that overexpress HOTTIP should also be examined. To further explore the relationship between HOTTIP and HCC invasiveness, HOTTIP detection in CTC from HCC patients will be conducted in our center.
In conclusion, the cancer transcriptome is far more complex than previously described. LncRNAs are believed to play a role in tumorigenesis and cancer development. The present study described the lncRNA HOTTIP as a valuable biomarker for the prediction of tumor recurrence in HCC patients after LT. We consider this characterization of HOTTIP to be an important part of our efforts to establish a prognostic model of recurrence and death that makes greater use of molecular markers in HCC patients after LT. Future laboratory work will also focus on the exact mechanisms by which HOTTIP induces malignant biological behavior in HCC cells.
Acknowledgements
Funding: This study was supported by the Key Research Development Program of Zhejiang Province (No. 2018C03018), the Technology Research Program of Jiaxing (Grant No. 2015AY23013).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: This study was approved by the Medical Ethics Committee of the First Affiliated Hospital of Zhejiang University (NO. 2016209), and informed consent was obtained from all patients.
References
- Jemal A, Bray F, Center MM, et al. Global cancer statistics. CA Cancer J Clin 2011;61:69-90. [Crossref] [PubMed]
- El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology 2007;132:2557-76. [Crossref] [PubMed]
- Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015;136:E359-86. [Crossref] [PubMed]
- Dutkowski P, Linecker M, DeOliveira ML, et al. Challenges to liver transplantation and strategies to improve outcomes. Gastroenterology 2015;148:307-23. [Crossref] [PubMed]
- Ramesh H. Resection for hepatocellular carcinoma. J Clin Exp Hepatol 2014;4:S90-6. [Crossref] [PubMed]
- Arzumanyan A, Reis HM, Feitelson MA. Pathogenic mechanisms in HBV- and HCV-associated hepatocellular carcinoma. Nat Rev Cancer 2013;13:123-35. [Crossref] [PubMed]
- Guttman M, Amit I, Garber M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 2009;458:223-7. [Crossref] [PubMed]
- Dinger ME, Amaral PP, Mercer TR, et al. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res 2008;18:1433-45. [Crossref] [PubMed]
- Wang KC, Yang YW, Liu B, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 2011;472:120-4. [Crossref] [PubMed]
- Cillo C, Cantile M, Faiella A, et al. Homeobox genes in normal and malignant cells. J Cell Physiol 2001;188:161-9. [Crossref] [PubMed]
- Cillo C, Schiavo G, Cantile M, et al. The HOX gene network in hepatocellular carcinoma. Int J Cancer 2011;129:2577-87. [Crossref] [PubMed]
- Quagliata L, Matter MS, Piscuoglio S, et al. Long noncoding RNA HOTTIP/HOXA13 expression is associated with disease progression and predicts outcome in hepatocellular carcinoma patients. Hepatology 2014;59:911-23. [Crossref] [PubMed]
- Tsang FH, Au SL, Wei L, et al. Long non-coding RNA HOTTIP is frequently up-regulated in hepatocellular carcinoma and is targeted by tumour suppressive miR-125b. Liver Int 2015;35:1597-606. [Crossref] [PubMed]
- Ge Y, Yan X, Jin Y, et al. MiRNA-192 [corrected] and miRNA-204 Directly Suppress lncRNA HOTTIP and Interrupt GLS1-Mediated Glutaminolysis in Hepatocellular Carcinoma. PLoS Genet 2015;11:e1005726. [Crossref] [PubMed]
- Li Z, Zhao X, Zhou Y, et al. The long non-coding RNA HOTTIP promotes progression and gemcitabine resistance by regulating HOXA13 in pancreatic cancer. J Transl Med 2015;13:84. [Crossref] [PubMed]
- Zheng SS, Xu X, Wu J, et al. Liver transplantation for hepatocellular carcinoma: Hangzhou experiences. Transplantation 2008;85:1726-32. [Crossref] [PubMed]
- Xu X, Lu D, Ling Q, et al. Liver transplantation for hepatocellular carcinoma beyond the Milan criteria. Gut 2016;65:1035-41. [Crossref] [PubMed]
- Zhang F, Wu LM, Zhou L, et al. Predictive value of expression and promoter hypermethylation of XAF1 in hepatitis B virus-associated hepatocellular carcinoma treated with transplantation. Ann Surg Oncol 2008;15:3494-502. [Crossref] [PubMed]
- Wu LM, Zhang F, Xie HY, et al. MMP2 promoter polymorphism (C-1306T) and risk of recurrence in patients with hepatocellular carcinoma after transplantation. Clin Genet 2008;73:273-8. [Crossref] [PubMed]
- Mazzaferro V, Llovet JM, Miceli R, et al. Predicting survival after liver transplantation in patients with hepatocellular carcinoma beyond the Milan criteria: a retrospective, exploratory analysis. Lancet Oncol 2009;10:35-43. [Crossref] [PubMed]
- Zimmerman MA, Ghobrial RM, Tong MJ, et al. Recurrence of hepatocellular carcinoma following liver transplantation: a review of preoperative and postoperative prognostic indicators. Arch Surg 2008;143:182-8; discussion 188. [Crossref] [PubMed]
- Schutte K, Schulz C, Link A, et al. Current biomarkers for hepatocellular carcinoma: Surveillance, diagnosis and prediction of prognosis. World J Hepatol 2015;7:139-49. [Crossref] [PubMed]
- Yao FY, Ferrell L, Bass NM, et al. Liver transplantation for hepatocellular carcinoma: expansion of the tumor size limits does not adversely impact survival. Hepatology 2001;33:1394-403. [Crossref] [PubMed]
- Yang Z, Zhou L, Wu LM, et al. Overexpression of long non-coding RNA HOTAIR predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation. Ann Surg Oncol 2011;18:1243-50. [Crossref] [PubMed]
- Schwartz M, Dvorchik I, Roayaie S, et al. Liver transplantation for hepatocellular carcinoma: extension of indications based on molecular markers. J Hepatol 2008;49:581-8. [Crossref] [PubMed]
- Schmidt C, Marsh JW. Molecular signature for HCC: role in predicting outcomes after liver transplant and selection for potential adjuvant treatment. Curr Opin Organ Transplant 2010;15:277-82. [Crossref] [PubMed]
- Wang FS, Fan JG, Zhang Z, et al. The global burden of liver disease: the major impact of China. Hepatology 2014;60:2099-108. [Crossref] [PubMed]
- Kim JH, Sohn BH, Lee HS, et al. Genomic predictors for recurrence patterns of hepatocellular carcinoma: model derivation and validation. PLoS Med 2014;11:e1001770. [Crossref] [PubMed]


