
Detecting donor-specific antibodies: the importance of sorting the wheat from the chaff
Introduction
Immunological injury to the allograft remains one of the major limitations to graft survival after solid organ transplantation and there is overwhelming evidence which implicates antibodies to donor specific HLA (human leukocyte antigen) in this process for kidney, pancreas, heart, lung, and small bowl transplant (1-4). There are also emerging but controversial evidences for HLA antibodies in liver transplantation (5-7). The existence of HLA was established in the 1950s following the observation that sera from individuals who had previously had a blood transfusion or pregnancy caused the agglutination of donor lymphocytes (8). Hyperacute rejection of a renal allograft mediated by pre-existing donor-specific HLA antibodies (DSA) was first described in 1966 (9) and the prognostic implications of a positive cytotoxic cross match between recipient’s serum and donor’s lymphocytes was subsequently recognised in Patel and Terasaki’s ground breaking research (2). The ability to determine the existence of preformed DSA by cross match techniques and to avoid transplantation when these were present resulted in the elimination of hyperacute rejection and a significant reduction in the incidence of acute accelerated rejection (2,10,11). These initial developments in HLA were of fundamental importance in establishing satisfactory outcomes after solid organ transplantation and promoting its widespread acceptance as a treatment. There have been many advances since. In 1964, the International Histocompatibility Workshops were established and these have facilitated global collaboration between HLA laboratories. This has been key to the development of serological, and later genetic, methods of determining an individual’s HLA type and to the optimisation of assays for detection for HLA antibodies, most recently including Luminex technology. Today, medium resolution genotyping of the donor and recipient is common practice in solid organ transplantation and Luminex assays, which provide semi-quantitative information on HLA antibodies, are widely used to inform clinical decision making (3). These more recent advances have stimulated the growth of HLA incompatible transplantation programmes; something that would have been unthinkable 60 years ago.
This article focuses on the role of HLA in alloimmune injury in transplantation and the relative merits and disadvantages of the available laboratory methods for detecting DSA.
What are HLA?
HLA are glycoproteins which are expressed on the surface of all nucleated cells; their primary role is to bind peptides and present them to T cell receptors. In this capacity, HLA are essential to the development of an individual’s T cell repertoire and to protection against infection and malignancy (12).
HLA can be divided into the classical class I antigens (HLA-A, B and C) which are present on all nucleated cells and class II antigens (HLA-DR, DP and DQ) which exist on antigen presenting cells only. Class I HLA are formed from a polymorphic α polypeptide chain with a constant β2 microglobulin subunit and are assembled by chaperone proteins in the endoplasmic reticulum; class I HLA present peptides of intracellular origin (which have been loaded from the cytoplasm) to the receptors of CD8+ T cells (Figure 1A). Class II HLAs are constitutively expressed on B cells and other antigen presenting cells, although expression can also be induced on other cells such as the endothelial cells and activated T cells in the context of inflammation (13). HLA class II are comprised of a constant α and polymorphic β chain for DR and polymorphic α and β chains for DQ and DP. Class II HLAs present peptides of extracellular origin (which are loaded from the cell’s phagolysosome) to the receptors of CD4+ T cells (Figure 1B) (12).
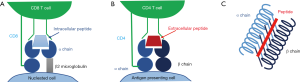
In both class I and class II HLA, peptides are presented within a peptide binding groove which is composed of “walls” of two alpha helices and a beta pleated sheath “floor” (Figure 1C). The amino acid sequence in this part of the HLA is highly polymorphic and determines its peptide binding repertoire. It is this antigen presenting region-peptide complex that interacts with the T cell receptor.
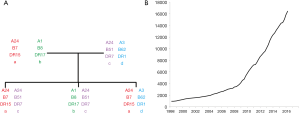
The genes which encode HLA are found in the major histocompatibility complex (MHC) on the short arm of chromosome 6 and demonstrate codominance in their inheritance (Figure 2). In addition to the classical HLA genes (HLA-A, B, C, DR, DP, DQ), non-classical HLA genes and other genes involved in immunity are also present in the MHC region (14). Unlike most genes in the human genome which have two or three major alleles, the HLA genes are highly polymorphic with over 16,000 HLA alleles identified to date; this number is increasing exponentially with advances in HLA typing (Figure 2). The polymorphic nature of HLA is likely to confer a population survival benefit by maximising the diversity of non-self peptides which can be presented to T cells and inducing robust immunological responses to infection. Some HLA types have been associated with protection against infections such as HIV (15,16). However, this polymorphism in HLA is a major barrier for transplantation as HLA mismatches between recipients and unrelated donors are very common.
How do HLA antibodies develop?
The exposure of an individual’s immune system to non-self HLA may result in the generation of HLA antibodies; this usually occurs via three mechanisms: transfusion, transplantation and pregnancy. The vast degree of polymorphism in the HLA system results in a large number of non-self stimuli for antibody development.
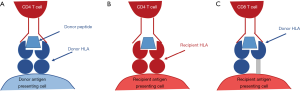
The initial step in immune recognition of non-self HLA is allorecognition. It is hypothesised that there are three pathways of allorecognition: the direct, indirect and semi-direct pathways (17) (Figure 3). In antibody generation, the indirect pathway is of particular importance because of its role in the generation of allo-specific CD4+ T cells which facilitate class-switching of short-lived IgM-producing B cell into long-lived IgG producing B cells. In this allorecognition pathway, an individual’s antigen presenting cells present fragments of endocytosed non-self HLA on a self HLA class II antigen. In the context of the appropriate costimulatory signals, the cognate CD4+ T cell is activated and may differentiate into a TFH cell which provides help to B cells in the process of activation and differentiation (17-19).
B cells are exposed to antigens presented by follicular dendritic cells within the secondary lymphoid tissues. When a B cell receptor binds to its cognate antigen, costimulatory signalling stimulates receptor-mediated endocytosis of the antigen which is processed and presented on the B cell’s HLA class II. The activated B cell migrates to the border of the B cell/T cell zone within the secondary lymphoid tissues to seek the help from a complementary TFH cell. The B cell and TFH cell form a germinal centre in which B cell proliferation and differentiation into plasma cells occurs. In this process, the specificity of the antibody to the antigen is enhanced (a process known as somatic hypermutation) and antibody class switching occurs; this usually results in the formation of plasma cells which generate antibody with very high affinity for the non-self antigen. In addition, this process results in the formation of long lived plasma cells, which can reside in the bone marrow and generate HLA antibodies for many years, and B memory cells, which can circulate in a quiescent state for decades but remain readily activated by re-exposure to the stimulating HLA antigen (20,21).
In the earliest days of HLA science, it was recognised that HLA antibodies generated following exposure to a single non-self HLA antigen were cross reactive with other non-self HLA which appeared unrelated to the immunising antigen e.g., individuals sensitized to HLA-A2 developed an antibody which also reacted with HLA-B57 and B58. These groups of antigens which were cross reactive with a single antibody became known as the cross reactive groups (CREGs) (22). It was hypothesised that these antigens shared “determinants” which were key to the specificity of the antibody; these determinants latterly became known as epitopes (23). Epitopes are the part of an antigen which is initially recognised by the B cell receptor and to which an antibody binds. An eplet is a 3 Å area on the antigen surface comprising a small number of amino acids which exist in close proximity in the tertiary HLA structure, and can be predicted by HLA matchmaker (24). Most of the hypothetical eplets are concordant with known CREGs, Terasaki epitopes (25) identified by serological adsorption/elution studies (where antibodies are eluted from one HLA antigen and then adsorption by another HLA antigen with a shared epitope is demonstrated) and antibody profiles from single antigen bead assays (26). For example, cross reactivity between A2, B57 B58, can be explained by well known subgroup of A2 CREG, called 17P which recognises GETERK (amino acid position 62–66) epitope (22), 62GE eplets by HLA matchmaker (24), and 62G by Terasaki epitopes (25).
What are the consequences of DSA in transplantation?
DSA in the recipient serum may exist prior to transplantation (pre-formed DSA) or develop as a consequence of sensitisation to the mismatched donor HLA antigens (de novo DSA). DSA binding to donor HLA on the endothelial surface has a number of potential consequences.
Complement activation
The complement fixing capacity of DSA is determined by the antibody class; the majority of DSA detected in transplantation are IgG or IgM which are both potentially complement fixing. Within the IgG class, antibody subclass determines the capacity to fix complement with IgG3 and IgG1 being potent activators of the complement cascade (27). Complement fixing antibodies bind to the graft endothelium resulting in initiation of the classical complement pathway (28). This process results in the generation of products which recruit inflammatory cells into the graft, opsonise the donor endothelial cells making them targets for neutrophils and macrophages and stimulate cytokine synthesis resulting in vasodilation and leucocyte extravasation into the transplanted organ (28,29). The membrane attack complex is the final product of the complement cascade and results in direct lysis of the antibody-coated cells (30). The presence of complement fixing DSA in solid organ transplantation has traditionally been demonstrated by performing immunofluorescence for C4d, a by-product of the classical complement pathway, on allograft biopsies.
Antibody dependent cell mediated cytotoxicity (ADCC)
When DSA bind to the graft endothelium, the crystalline fragment (Fc) of the bound antibody can act as a stimulus to innate immune cells. FcƔ receptors (FcƔRs) are activatory receptors for neutrophils and macrophages and the most potent stimulus of natural killer cell (NKC) activation. The interaction between an antibody’s Fc and the FcƔRIIIa on the NKC results in the formation of a synapse across which the NKC secretes perforins and granzymes resulting in apoptosis of the target cell. This interaction also stimulates the generation of chemokines and cytokines which enhance HLA expression on the donor endothelium and recruit inflammatory cells (31,32). Both complement-fixing IgG1/3, and IgG2 or IgG4 DSAs which are not good at fixing complement, can induce ADCC. The microvascular inflammation present in allografts in the presence of DSA but the absence of C4d deposition is believed to be predominantly driven by NKC-mediated antibody dependent cell mediated cytotoxicity (31-33).
Modification of the vascular endothelium
There is emerging evidence that DSA binding to HLA, particularly HLA class I, on the vascular endothelium initiates an intracellular signalling cascade with implications for endothelial cell structure and function. These modifications include increased expression of leucocyte adhesion ligands, alteration of the cytoskeleton and enhanced cell proliferation and survival (34). These changes contribute to the classical histological features of fibrosis and intimal proliferation which is characteristic of chronic antibody mediated rejection in all solid organ transplants (35,36).
Accommodation
DSA have the potential to induce allograft damage by any of the mechanisms described but there is a cohort of patients with detectable DSA but no histological evidence of inflammation or allograft damage (37). In these cases, the graft appears to have “accommodated” the antibodies without a detrimental effect, especially in liver transplantation, or ABO-incompatible organ transplantation. The physiology of this is poorly understood.
How are DSA detected in the HLA laboratory?
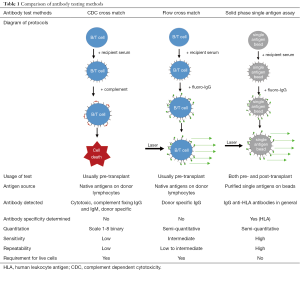
The accurate detection of pre-existing donor specific antibodies in the laboratory is of fundamental importance in determining the immunological risk associated with transplanting a particular organ (3). Traditionally, donor specific antibodies have been detected at the time of transplantation by performing a cross match (2). The complement dependent cytotoxicity (CDC) cross match is the oldest test in the HLA laboratory and involves extracting donor lymphocytes from blood or lymphoid tissue, incubating donor cells with recipient serum followed by rabbit complement and adding dyes to distinguish dead from living donor cells. This process detects the presence of antibody-antigen interaction on cell surface which activates complement and cause cell death. The flow cross match similarly involves the incubation of donor cells with recipient serum but, instead of complement, a fluorochrome-labelled anti-human IgG is added. This detector antibody will bind to antibodies which have been bound on the donor cell surface. In addition, fluorescent labelled antibodies specific to B and T lymphocytes are added to the donor cells. In a flow cytometer, laser excitation identifies the lymphocytes and the presence of the detector antibody on the cell surface; this correlates with the quantity of bound antibodies. Cut-off for positive flow cross match is usually determined by how much is the normal variance (or standard deviation, SD) of the flow crossmatches with negative control sera without known anti-HLA antibodies. If the fluorescent with serum from tested patient is 2SD or 3SD stronger than that with negative controls, the flow cross match will be called as positive. So positive flow cross match is determined statistically and might not be biologically relevant. In most cases a positive cross match, either with CDC or flow cross match, indicates DSA binding to the donor cells. This is not always true as auto antibodies or unknown non-HLA factors might cause a false positive cross match. The specificity of DSA cannot be easily determined using cross match assays because usually more than one HLA are expressed on donor cells.
Solid phase technology differs from the cross match tests because the HLA source is manufactured beads coated with multiple HLA class I or II antigens (Phenotype beads or PRA beads) or a single HLA antigen [single antigen beads (SAB)]. HLA antibody testing using solid phase assays involves incubating the beads with the recipient serum and adding a fluorochrome-labelled anti-human IgG secondary antibody. The fluorescent signal can be detected using a flow cytometer, or more commonly, a Luminex analyser. In the latter case, one laser determines the bead identity by its emission of light of two unique wavelengths while another identifies the intensity of fluorochrome on the bead. This is a semi-quantitative test which identifies the presence, the relative strength and the specificity of HLA antibodies. Luminex technology has a number of advantages over cross matching for the detection of antibodies, most notably the high sensitivity, good repeatability, ability to easily determine HLA antibody specificity, and independency of living cells.
Table 1 describes the laboratory differences between the antibody testing methods. There are also differences in the clinical relevance of the antibodies detected by these methods in transplantation. CDC cross match positivity has been associated with hyperacute and acute accelerated rejection in all solid organ transplants (2,38,39) while the association between a positive flow cross match and poor allograft outcomes is less robust (40). Pre-existing DSA identified by the Luminex single antigen assay alone have been associated with an increased risk of rejection and allograft loss in kidney transplantation, but this effect is less clear in other solid organ transplants (41). In each of these tests, there is a trade-off between specificity and sensitivity. The immunological risk for a particular transplant must assessed with multiple methods and also be balanced against the recipient’s risk of not being transplanted; this necessitates a personalised decision-making process for each transplant recipient with input from the clinicians, the HLA laboratory and often the patient themselves.
Full table
What are single antigen bead assays and what are their limitations?
Luminex SAB technology has transformed antibody assessment in transplantation by allowing the rapid determination of HLA antibody specificities at any time point in a high volume of samples. This has facilitated development of virtual cross matching (when the cross match result can be predicted from the recipient’s HLA antibodies if donor complete typings for HLA-A, B, C, DRB1, DRB3/4/5, DQA1/DQB1, DPA1/DPB1 are available), and made accurate DSA monitoring possible after transplantation (3).
Each class I or class II HLA SAB assay consists of 98 beads which are each coated with a single HLA antigen that has been stripped from a cell; the presence of HLA antibody is determined as described in Table 1. In addition to detecting the fluorescence emitted from the detector antibody, each bead emits a unique fluorescence which allows the HLA antigen specificity of the bead to be identified. The results are reported as a median fluorescent intensity (MFI). Luminex SAB is not a quantitative assay according to venders. The coefficient of variance for MFI could be up to 20–30% intra- or inter-labs. The variance can be reduced with standardization of testing protocols and diligent training of testing personals (42). However, the vender vs. vender, and lot vs. lot variances will not be resolved easily. To fully understand the utility of SAB in transplantation, it is necessary to have knowledge of its limitations:
Panel representation
Each class I and class II SAB assay contains 98 beads with 98 distinct HLA; this includes the HLA encoded by different alleles of the same antigen where the HLA molecules differ by at least one amino acid, e.g., A*02:01, A*02:03 and A*02:06. The antigens representation include most but NOT 100% common HLA antigens and a number of rarer HLA antigens (43). Nevertheless, given the huge diversity in HLA, it remains possible that HLA from a donor of ethnical minority might not be represented on a SAB kit so the presence of DSA cannot be definitively determined. However, due to cross reactivity among HLA antibodies, an antibody to a non-tested rare allele can usually be predicted with tested antigens sharing the same epitopes. This is very useful for HLA-DPB1 which has four distinct epitope groups (44). For example, non-tested antigens DPB1*02:02, 24, 40 share p56A & p85-87GPM epitopes with positive beads of DPB1*04:01, 15, 23, so they can be predicted to be positive if there are such antigens in the panel.
Bead saturation
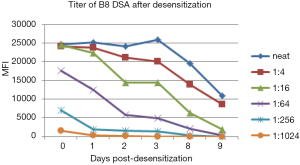
There are a limited number of HLA antigens on each bead in the SAB assay. Depending on instrumentation, the maximum MFI can be read out in Luminex is also limited. High titre HLA antibodies may saturate all of these targets, and make it difficult to quantify the true “strength” of the antibody present. In these cases, serial dilutions of the recipient serum do not result in a reduction in the MFI. For example in Figure 4, at day 1 after desensitization of a liver-kidney combined transplantation, B8 DSA titre dropped 4 times, from positive at 1:1,024 to positive at 1:256, but there is no difference in MFI of B8 DSA in neat serum because the strong DSA already saturated the beads.
Antigen density
The amount of antibody which is bound to each SAB depends not only upon how much antibody is present in the recipient serum but also upon how much target for that antibody exists. There is variation in the HLA antigen density between beads in the same kit, between assays from different manufacturers and between beads (used in the solid phase assay) and donor cells (used in the crossmatch) (3). Importantly, the amount of antigens coated on different beads in a SAB panel could differ twice, so a universal cut-off for all beads is impossible. PRA bead or phenotype bead, which has multiple HLA on the each bead, has much less antigen per HLA than SAB so the sensitivity to detect a specific antibody is much lower than SAB. The antigen density determines sensitivity and clinical relevance of an assay. For example, flow cross match positivity due to HLA-C, DP DSA is associated with antibody present at MFIs higher than flow cross match positivity attributed to DSA to other HLA loci (45). This reflects the reduced level of expression of HLA-C and HLA-DP on the cell surface (46). Variation in antigen density is one explanation for the imperfect correlation between DSA detected by SAB and cross match results (47).
False negativity for antibodies specific for shared epitopes
Many HLA antibodies are specific to public epitopes shared by many HLA rather than private epitopes on single HLA. When a single epitope is shared by multiple antigens, the complementary antibody will be spread over a large number of beads on the SAB assay. In consequence, there is less antibody binding to a single bead. As indicated in Figure 5A, MFIs for beads coated with DR8, DR11, DR12, DR13, DR14, DR17, DR18 are in the range of 160–500. However flow cross match with the patient’s daughter in presence of DR13 DSA, was borderline positive for B cells which express class II antigens. Usually weak DSA with MFI of 500 will not be able to cause positive B-cell flow cross match. However, the antibody binds to epitope p9EYST which is shared by these groups of HLA-DR8, DR11, DR12, DR13, DR14, DR17, DR18. If there is only a single DR13 bead in the panel, the MFI to DR13 bead would be much higher. So when donors cells with only DR13 antigen were used in the flow cross match, the antibody bind significantly more positive than negative control sera. The antibodies are certainly real because this female patient (self-antigen DR4, DR15) was clearly sensitized in pregnancy to paternal mismatched antigen DR13. The authenticity of these antibodies was validated 3 month later: much stronger antibodies (MFI up to 10,000) were found when an infection breakout recalled memory B cell responses to epitope p9EYST (Figure 5B). This phenomenon is common for HLA antigens which share a “public” epitope such as Bw4, Bw6, and DR52 group (DR8, DR11, DR12, DR13, DR14, DR17, and DR18) but can also be seen for other CREGs. Donors with these broad antigens are very common [Bw4 (74%), Bw6 (85%), DR52 group (68%)] so the problem will not be in rarity. If this is not recognised, it is possible to fail to identify a real HLA antibody which is actually present at higher titre than what was indicated on the SAB and could result in early accelerated antibody mediated rejection.

False negativity due to prozone
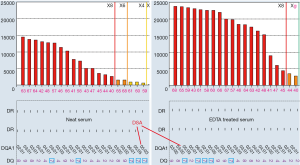
There are a number of instances where endogenous substances in the recipient serum may interfere with the detection of HLA antibodies leading to false negativity; this has been attributed to the C1 complex, IgM antibodies and high titre IgG antibodies (48-50) and is known as the prozone effect. Laboratories employ methods such as the addition of EDTA, heat inactivation, addition of DTT (Dithiothreitol) and serial dilutions to overcome this. Figure 6 illustrated that EDTA removed prozone and uncovered strong DSA to DQ7 in a patient who experienced antibody-mediated rejection after kidney transplantation. In neat serum without EDTA treatment, MFI for DSA to DQ7 (paired with DQA1*05) is less than 1,000 and would be called as negative in many labs, and is not consistent with the presence of active AMR. However, the true MFI of the DSA was in fact more than 20,000 as indicated in EDTA-treated serum. Prozone effect is more often to be seen in sensitized patients who are previously transplanted. EDTA treatment seems to be the easiest, most cost-effective approach to remove prozone and has been routinely used by all HLA labs in Canada. Recent study (50) concluded serial dilution is the only solution for prozone. However, it is unclear in the study, EDTA was used for all sera, or the concentration and source of EDTA (51). Whether prozone effect after proper EDTA treatment still exists or not deserves more investigations.
False positivity due to antigen configuration or other unknown factors
The process of purifying HLA antigens from cells and conjugating them to the beads can result in distortion of the 3-dimensional structure for HLA. Antibodies to denatured beta-2-microglobulin free HLA class I on SAB might not be detrimental for organ transplant (52). As a consequence, non-native epitopes may be exposed in the SAB assay which are not accessible to antibodies in vivo and vice versa. This may result in false positivity when antibodies are detected to epitopes that are not exposed in vivo or false negativity if an antibody is present to an epitope that is exposed on the cell surface but becomes distorted in the manufacturing process. In studies of non-sensitized male patients, possible SAB false positivity has been detected for a number of HLA antigens (53).
In addition, exogenous substances such as intravenous immunoglobulin and monoclonal antibodies have also been reported to interfere with SAB test results (3) and interference from unknown factors can give a false positive result. This is usually (but not always) indicated by a high MFI value for the negative control bead in the SAB assay. Treatment with DTT, or absorption with naked uncoated beads, or filtration with NanoSep Columns, or foetal calf serum treatment have been used in labs to reduce false positivity. Unfortunately, there is no prefect solution for this problem because there are many different reasons for high background and many more are unknown.
Due to these limitations of the SAB assay, guideline, rather “cut off” might be better used to describe how a positive anti-HLA antibody is determined. It can be challenging to use single thresholds for positivity, for all beads in one SAB panel, for different sera, for different clinical applications. Ideally individual laboratories should validate their own threshold against cross match results and clinical outcomes.
How can it be determined if HLA antibodies are “real”?
In most transplant programmes, HLA antibody testing of waiting list individuals is performed at regular intervals prior to transplantation by Luminex SAB assays (3). Accurate determination of which detected HLA antibodies are relevant to transplantation is essential. Most solid organ transplant programmes (with the exception of some liver programmes) list HLA to which there are complementary antibodies in the patient’s serum as unacceptable donor mismatches (3,54). The purpose of this is to facilitate organ allocation and avoid organ transplantation with increased immunological risk. A failure to identify false positive antibodies from the Luminex assay results in the associated HLA being listed as unacceptable mismatches and unnecessarily limits the patient’s access to transplantation or results in the administration of unnecessary and costly treatments. This results in an increased risk of death on the waiting list, inequity for access to transplantation and potential adverse effects of enhanced immunosuppression (55-57). On the other hand, a failure to identify relevant HLA antibodies (false negatives) may result in the transplantation of organs with which there is an unanticipated increased risk of immunological injury that may prove detrimental to graft and recipient outcomes (38,41).
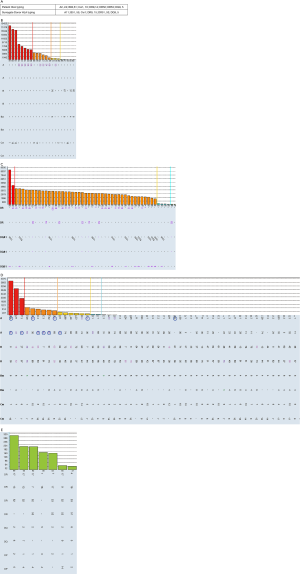
Similar to sorting wheat from chaff, distinguishing real antibodies from unreal “junk” reactivity in SAB can be very challenging. Here we use a case of false positive antibodies to explain common strategies to determine if HLA antibodies are real or not. Sera from a patient waiting for kidney transplantation were tested for anti-HLA antibodies using Luminex SAB (Figure S1). Class II SAB test (Figure S1C) identified HLA antibodies to all HLA-DR antigens (MFI =2,000–5,000) including self DR9, DR14, DR52, DR53. In class I SAB, strong antibodies to Cw12, Cw15 and self-antigen Cw1 (MFI =18,000), and moderate antibodies (MFI =1,000–8,000) to A10 CREG (A25,26,33,34,66,68,69) were identified (Figure S1B). The MFIs for negative control beads were in acceptable range (<400). Antibody profiles for repeating tests with DTT treatment, NanoSep Column and uncoated bead (Adsorb Out™) were as same as that from neat serum. The positive profile of Cw1, Cw12, and Cw12 can NOT be explained by extensive epitope analysis. High resolution typing for patient HLA-Cw1 was done and found patient self is C*01:02, which is the same antigen on the Cw1 bead. An individual cannot generate antibodies specific to own HLA type. It is puzzling to see such broad antibody reactivity in 20-year-old male without known sensitization events. Flow cross match with surrogate cells with DSA to Cw1, DR9, DR15, DR51, DR53 were cleanly negative. The same serum was tested again using Luminex PRA bead which demonstrates the complete absence of any class II HLA antibodies (Figure S1E) and lacking of positivity for beads carrying Cw1, CW12, and Cw15 (Figure S1D). Interestingly, beads coated with A10 CREG antigens, which are highlighted in circle (Figure S1D), were convincingly positive in PRA beads. Class II flow PRA beads were clearly negative too (data not shown). Positivity on bead coated with Cw1, Cw12, Cw15 but not A10 CREG antigens can be absorbed by non-relevant Cw17 single bead and bind more on beads with acid treated denatured antigens (data not shown). This indicated: (I) false positive Cw1, Cw12, and Cw15 beads are likely caused by denatured antigens; (II) reactivity to A10 CREG are likely to be real antibodies; (III) comparing with SAB, the less sensitive PRA beads might have more native antigens and be less prone to false positivity.
As highlighted in the case (Figure S1), multiple approaches shall be used to accurately determine what “real” anti-HLA antibodies are. These stratagems are summarized as below:
- Patients will not have antibodies to their self-antigens. Sometimes high resolution typing need to be done to determine what is real self-antigen (Figure S1). There are antibodies to a specific allele which belongs to a same antigen as the patient self-allele. For example, a patient self HLA is DPB1*04:01 could have strong antibodies to DPB1*04:02. These two DPB1*04 alleles have district and immunogenic epitopes which differ at amino acid position 56: A (Alanine) in DPB1*04:01 vs. E (Glutamic Acid) in DPB1*04:02. Other examples are A*66:02 vs. A*66:01, B*27:05 vs A*27:08, DRB1*04:02 vs. DRB1*04:01/04/05, DPB1*02:01 vs. DPB1*02:02. However, many of the allele-antibodies identified in SAB alone are not real antibodies, especially in non-sensitized patients. Epitope analysis and/or alternative testing methods are better to be done to distinguish real ones from unreal.
- Antibodies usually target for immunizing HLA or related cross reactive antigens. We need to know patient’s sensitization history including previous transplant (s) (mismatched donor HLA antigens), pregnancies (mismatched paternal HLA antigens, if known) and transfusions. Patients who were previously sensitized but recently quiescent, surgery, such as nephrectomy, or infection can boost amnestic memory responses and recall strong and broad anti-HLA antibodies [(58) and Figure 5]. Weaning immunosuppression alone, in fact, is a very prominent sensitization event (59).
- Using knowledge of epitope or CREGs to analyse SAB results is very important to make sense for specificities of the antibodies. The conventional CREG and CREG subgroups (22) can explain many antibody profiles for HLA-A, and B, but less for Cw, and class II antigens. There are pre-built convenient tools for CREG and/or epitope analysis in the antibody analysis softwares from both vendors of solid phase assays. Most of HLA class I and class II epitopes are covered in vender’s antibody analysis softwares. Another approach is to align and compare amino acid sequences for tested HLA allels in SAB panel, either manually, or using HLA Matchmaker software. HLA Matchmaker postulates HLA epitopes based on the amino acid sequences, known structures of some HLA antigens (60). More user-friendly tools for epitope analysis are still need to be developed. Epitope analysis is especially important for cases of weak antibodies (Figure 5) or seemingly unreal antibodies (Figure S1).
- There is no single assay prefect enough so multiple imperfect assays shall be used to determine true antibodies. These include but not limited to: more than one solid phase assay (i.e., SAB, phenotype beads and/or PRA screen beads) and/or SAB assay kits from more than one vender, one more than one platform (flow and Luminex), surrogate flow cross matches, acid treatment to denature antigens on beads, different methods for serum treatment such as EDTA, DTT, dilution, etc.
- It is good idea for labs to develop strategies for identifying and recognizing non-specific reactivity patterns, including “hot beads”, either by determining in-house or by published references. However, caution shall be taken because many well-established “hot” beads on which false positivity were found in many sera might give true antibodies in other sera: One man’s trash could be another man’s treasure!
What other factors may influence the pathogenicity of HLA DSA?
There are two questions that need to be answered regarding DSA: (I) is the DSA present in a patient or not? (II) Is the DSA clinically relevant or not for specific organ transplantation? SAB, although far from prefection, is the most powerful tool we ever had to answer 1st question. To answer the 2nd questions, there are other factors which have been hypothesised to impact the pathogenicity of donor specific HLA antibodies in solid organ transplantation.
The first factor is the expression of HLA on the allograft endothelium. Endothelial cells constitutively express class I HLA, with HLA-C expression being substantially less than HLA A and B, but expression of HLA class II can be induced in the context of inflammation, especially cell-mediated rejection (13). It is plausible that this may contribute to the increased incidence of acute rejection which is observed in kidney transplant recipients with delayed graft function. The aetiology of the delayed graft function, such as prolonged ischaemia, induces inflammation in the allograft which may increase the target density for HLA antibodies and alloreactive T cells (13). An effective method of determining HLA expression on the graft endothelium in real time is not yet available.
The second factor which may influence the pathogenicity of HLA antibodies is the avidity of the eplet-antibody interaction. The electrostatic potential of an amino acid polymorphism within an eplet influences the avidity of the antigen-antibody complex by affecting factors such as the formation of salt bridges and hydrogen bonds (61). Differences in electrostatic potential between polymorphic amino acids in the HLA class I of mismatched donor-recipient cross matches have previously been shown to correlate with both the presence and amount of donor specific class I HLA antibody (62).
The third factor which may affect the impact of HLA antibodies is their ability to fix complement. In the original CDC cross match, it was complement fixing, cytotoxic donor specific antibodies which were detected; this remains the test with greatest specificity for adverse transplant outcomes (38,40,63). In recent years, there has been interest in developing the Luminex SAB assay to detect not simply the presence of HLA antibodies but to determine those which can initiate the catabolism of complement. This is achieved by adding complement to the assay following incubation of the beads with recipient serum and then introducing a detector antibody which is specific for a product of the complement cascade (C1q or C3d) instead of for the human Fc receptor. A number of studies have identified an association between complement fixing antibodies detected by this method and an increased risk of antibody mediated rejection (64-66). However, it is likely that the majority of complement fixation in these assays is simply determined by the amount of HLA antibody present (27).
The fourth factor which may be useful in determining the impact of HLA antibodies is their IgG subclass. It is now apparent that following activation, plasma cells initially generate IgG3 antibodies. With ongoing eplet stimulation, other subclasses of eplet-specific IgG are generated moving from IgG3 to IgG1, IgG2 and finally IgG4 (67). The presence of IgG4 is associated with a refined immunological response and a prolonged immunogenic stimulus; this subclass is commonly detected in women who have been sensitised by pregnancy (67). A small number of studies have explored the impact of HLA IgG subclass in kidney transplantation and found an association between IgG1 and IgG3 and acute antibody mediated rejection while IgG4 is associated with chronic alloimmune injury (68). Pre-formed or de novo IgG3 DSA were also found to be the most detrimental type of DSA in survival of liver transplantation (7).
It is biologically plausible that each of these factors is related to the pathogenicity and potential impact of HLA antibodies and evidence is accumulating that an association exists between some of these and transplant outcomes. However, it is not yet clear how this could be incorporated into clinical practice to further optimise solid organ transplant outcomes without unnecessarily restricting the access of potential recipients to transplantation.
Once DSA were detected either pre-transplantation or post-transplantation, they must be respected. This is true more for transplantations of kidney, pancreas, heart, lung grafts, less for liver, small bowel grafts. The best approach of handing pre-transplant DSA is to avoid it. However, for patients with broader allo-sensitization, i.e., very high PRA, desensitization is necessary to increase accessibility for transplantation. Crossing pre-transplantation DSA might also be only option for medically urgent cases, especially for patients waiting for heart or lung transplants. Antibody-mediated rejection (AMR), either acute or chronic, caused by either preformed or de novo DSAs are detrimental for survival of organ transplant. To reduce the level of anti-HLA antibodies, many therapeutical regimens can be used. Usually they are combination of plasmapheresis (PP), immunoadsorption (IA), intravenous immunoglobulin (IVIg), rituximab, proteasome inhibitors such as bortezomib, complement inhibitors (69,70). Applications of these stratagems prior to kidney (69,71,72), heart (73-75) and lung (57) transplantation, or treatment of AMR (4,76), have been reviewed extensively in published literatures.
Conclusions
Graft damage from alloimmune injury and the reduced access of highly sensitised patients to transplantation are two of the major challenges facing the transplant community. The accurate determination of clinically relevant HLA antibodies and an appropriate interpretation of their impact are essential in addressing these issues. In the detection of HLA antibodies, a single perfect test providing the desirable accuracy, quantitation, sensitivity and specificity does not exist. As a consequence, HLA laboratories must incorporate multiple laboratory tests and analyses with information about each individual to ensure that the correct information regarding immunological risk is provided for each patient. Similar approaches were recommended in working group report of Sensitization in Transplantation: Assessment of Risk (STAR) (77).
Acknowledgements
None.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Safavi S, Robinson DR, Soresi S, et al. De novo donor HLA-specific antibodies predict development of bronchiolitis obliterans syndrome after lung transplantation. J Heart Lung Transplant 2014;33:1273-81. [Crossref] [PubMed]
- Patel R, Terasaki PI. Significance of the positive crossmatch test in kidney transplantation. N Engl J Med 1969;280:735-9. [Crossref] [PubMed]
- Tait BD, Susal C, Gebel HM, et al. Consensus guidelines on the testing and clinical management issues associated with HLA and non-HLA antibodies in transplantation. Transplantation 2013;95:19-47. [Crossref] [PubMed]
- Manfredini V, Leone O, Agostini V, et al. Antibody-mediated rejection in heart transplantation: new developments and old uncertainties. Curr Opin Organ Transplant 2017;22:207-14. [Crossref] [PubMed]
- O'Leary JG, Demetris AJ, Friedman LS, et al. The role of donor-specific HLA alloantibodies in liver transplantation. Am J Transplant 2014;14:779-87. [Crossref] [PubMed]
- Taner T, Stegall MD, Heimbach JK. Antibody-mediated rejection in liver transplantation: current controversies and future directions. Liver Transpl 2014;20:514-27. [Crossref] [PubMed]
- O'Leary JG, Kaneku H, Banuelos N, et al. Impact of IgG3 subclass and C1q-fixing donor-specific HLA alloantibodies on rejection and survival in liver transplantation. Am J Transplant 2015;15:1003-13. [Crossref] [PubMed]
- Dausset J. Iso-leuko-antibodies. Acta Haematol 1958;20:156-66. [Crossref] [PubMed]
- Kissmeyer-Nielsen F, Olsen S, Petersen VP, et al. Hyperacute rejection of kidney allografts, associated with pre-existing humoral antibodies against donor cells. Lancet 1966;2:662-5. [Crossref] [PubMed]
- Cook DJ, Terasaki PI, Iwaki Y, et al. The flow cytometry crossmatch in kidney transplantation. Clin Transpl 1987.409-14. [PubMed]
- Gebel HM, Bray RA, Nickerson P. Pre-transplant assessment of donor-reactive, HLA-specific antibodies in renal transplantation: contraindication vs. risk. Am J Transplant 2003;3:1488-500. [Crossref] [PubMed]
- Wieczorek M, Abualrous ET, Sticht J, et al. Major Histocompatibility Complex (MHC) Class I and MHC Class II Proteins: Conformational Plasticity in Antigen Presentation. Front Immunol 2017;8:292. [Crossref] [PubMed]
- Wedgwood JF, Hatam L, Bonagura VR. Effect of interferon-gamma and tumor necrosis factor on the expression of class I and class II major histocompatibility molecules by cultured human umbilical vein endothelial cells. Cell Immunol 1988;111:1-9. [Crossref] [PubMed]
- Neefjes J, Jongsma ML, Paul P, et al. Towards a systems understanding of MHC class I and MHC class II antigen presentation. Nat Rev Immunol 2011;11:823-36. [Crossref] [PubMed]
- International HIV Controllers Study, Pereyra F, Jia X, et al. The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science 2010;330:1551-7.
- Apps R, Qi Y, Carlson JM, et al. Influence of HLA-C expression level on HIV control. Science 2013;340:87-91. [Crossref] [PubMed]
- Ali JM, Bolton EM, Bradley JA, et al. Allorecognition pathways in transplant rejection and tolerance. Transplantation 2013;96:681-8. [Crossref] [PubMed]
- Conlon TM, Saeb-Parsy K, Cole JL, et al. Germinal center alloantibody responses are mediated exclusively by indirect-pathway CD4 T follicular helper cells. J Immunol 2012;188:2643-52. [Crossref] [PubMed]
- Taylor AL, Negus SL, Negus M, et al. Pathways of helper CD4 T cell allorecognition in generating alloantibody and CD8 T cell alloimmunity. Transplantation 2007;83:931-7. [Crossref] [PubMed]
- Clatworthy MR, Espeli M, Torpey N, et al. The generation and maintenance of serum alloantibody. Curr Opin Immunol 2010;22:669-81. [Crossref] [PubMed]
- Bayles I, Milcarek C. Plasma cell formation, secretion, and persistence: the short and the long of it. Crit Rev Immunol 2014;34:481-99. [Crossref] [PubMed]
- Rodey G. HLA beyond tears: introduction to human histocompatibility. 2nd ed. Pel-Freez; 2000.
- Schwartz BD, Luehrman LK, Rodey GE. Public antigenic determinant on a family of HLA-B molecules. J Clin Invest 1979;64:938-47. [Crossref] [PubMed]
- Duquesnoy RJ, Marrari M. Correlations between Terasaki's HLA class I epitopes and HLAMatchmaker-defined eplets on HLA-A, -B and -C antigens. Tissue Antigens 2009;74:117-33. [Crossref] [PubMed]
- El-Awar N, Lee JH, Tarsitani C, et al. HLA class I epitopes: recognition of binding sites by mAbs or eluted alloantibody confirmed with single recombinant antigens. Hum Immunol 2007;68:170-80. [Crossref] [PubMed]
- Marrari M, Duquesnoy RJ. Correlations between Terasaki's HLA class II epitopes and HLAMatchmaker-defined eplets on HLA-DR and -DQ antigens. Tissue Antigens 2009;74:134-46. [Crossref] [PubMed]
- Schaub S, Honger G, Koller MT, et al. Determinants of C1q binding in the single antigen bead assay. Transplantation 2014;98:387-93. [Crossref] [PubMed]
- Ehrnthaller C, Ignatius A, Gebhard F, et al. New insights of an old defense system: structure, function, and clinical relevance of the complement system. Mol Med 2011;17:317-29. [Crossref] [PubMed]
- Markiewski MM, Lambris JD. The role of complement in inflammatory diseases from behind the scenes into the spotlight. Am J Pathol 2007;171:715-27. [Crossref] [PubMed]
- Rother RP, Rollins SA, Mojcik CF, et al. Discovery and development of the complement inhibitor eculizumab for the treatment of paroxysmal nocturnal hemoglobinuria. Nat Biotechnol 2007;25:1256-64. [Crossref] [PubMed]
- Resch T, Fabritius C, Ebner S, et al. The Role of Natural Killer Cells in Humoral Rejection. Transplantation 2015;99:1335-40. [Crossref] [PubMed]
- Sis B, Halloran PF. Endothelial transcripts uncover a previously unknown phenotype: C4d-negative antibody-mediated rejection. Curr Opin Organ Transplant 2010;15:42-8. [Crossref] [PubMed]
- Lee CY, Lotfi-Emran S, Erdinc M, et al. The involvement of FcR mechanisms in antibody-mediated rejection. Transplantation 2007;84:1324-34. [Crossref] [PubMed]
- Valenzuela NM, Reed EF. Antibodies to HLA Molecules Mimic Agonistic Stimulation to Trigger Vascular Cell Changes and Induce Allograft Injury. Curr Transplant Rep 2015;2:222-32. [Crossref] [PubMed]
- Mehra MR, Crespo-Leiro MG, Dipchand A, et al. International Society for Heart and Lung Transplantation working formulation of a standardized nomenclature for cardiac allograft vasculopathy-2010. J Heart Lung Transplant 2010;29:717-27. [Crossref] [PubMed]
- Haas M, Sis B, Racusen LC, et al. Banff 2013 meeting report: inclusion of c4d-negative antibody-mediated rejection and antibody-associated arterial lesions. Am J Transplant 2014;14:272-83. [Crossref] [PubMed]
- Tang AH, Platt JL. Accommodation of grafts: implications for health and disease. Hum Immunol 2007;68:645-51. [Crossref] [PubMed]
- Opelz G, Dohler B, Susal C. Analysis of positive kidney, heart, and liver transplant crossmatches reported to the Collaborative Transplant Study. Hum Immunol 2009;70:627-30. [Crossref] [PubMed]
- Ruiz R, Tomiyama K, Campsen J, et al. Implications of a positive crossmatch in liver transplantation: a 20-year review. Liver Transpl 2012;18:455-60. [Crossref] [PubMed]
- Orandi BJ, Garonzik-Wang JM, Massie AB, et al. Quantifying the risk of incompatible kidney transplantation: a multicenter study. Am J Transplant 2014;14:1573-80. [Crossref] [PubMed]
- Mohan S, Palanisamy A, Tsapepas D, et al. Donor-specific antibodies adversely affect kidney allograft outcomes. J Am Soc Nephrol 2012;23:2061-71. [Crossref] [PubMed]
- Reed EF, Rao P, Zhang Z, et al. Comprehensive assessment and standardization of solid phase multiplex-bead arrays for the detection of antibodies to HLA-drilling down on key sources of variation. Am J Transplant 2013;13:3050-1. [Crossref] [PubMed]
- Pei R, Lee JH, Shih NJ, et al. Single human leukocyte antigen flow cytometry beads for accurate identification of human leukocyte antigen antibody specificities. Transplantation 2003;75:43-9. [Crossref] [PubMed]
- Cano P, Fernández-Viña M. Two sequence dimorphisms of DPB1 define the immunodominant serologic epitopes of HLA-DP. Hum Immunol 2009;70:836-43. [Crossref] [PubMed]
- Bachelet T, Martinez C, Del Bello A, et al. Deleterious Impact of Donor-Specific Anti-HLA Antibodies Toward HLA-Cw and HLA-DP in Kidney Transplantation. Transplantation 2016;100:159-66. [Crossref] [PubMed]
- Muczynski KA, Cotner T, Anderson SK. Unusual expression of human lymphocyte antigen class II in normal renal microvascular endothelium. Kidney Int 2001;59:488-97. [Crossref] [PubMed]
- Gloor JM, Winters JL, Cornell LD, et al. Baseline donor-specific antibody levels and outcomes in positive crossmatch kidney transplantation. Am J Transplant 2010;10:582-9. [Crossref] [PubMed]
- Weinstock C, Schnaidt M. The complement-mediated prozone effect in the Luminex single-antigen bead assay and its impact on HLA antibody determination in patient sera. Int J Immunogenet 2013;40:171-7. [Crossref] [PubMed]
- Kosmoliaptsis V, Bradley JA, Peacock S, et al. Detection of immunoglobulin G human leukocyte antigen-specific alloantibodies in renal transplant patients using single-antigen-beads is compromised by the presence of immunoglobulin M human leukocyte antigen-specific alloantibodies. Transplantation 2009;87:813-20. [Crossref] [PubMed]
- Tambur AR, Herrera ND, Haarberg KM, et al. Assessing Antibody Strength: Comparison of MFI, C1q, and Titer Information. Am J Transplant 2015;15:2421-30. [Crossref] [PubMed]
- Wiebe C, Gareau AJ, Pochinco D, et al. Evaluation of C1q Status and Titer of De Novo Donor-Specific Antibodies as Predictors of Allograft Survival. Am J Transplant 2017;17:703-11. [Crossref] [PubMed]
- Cai J, Terasaki PI, Anderson N, et al. Intact HLA not beta2m-free heavy chain-specific HLA class I antibodies are predictive of graft failure. Transplantation 2009;88:226-30. [Crossref] [PubMed]
- Morales-Buenrostro LE, Terasaki PI, Marino-Vazquez LA, et al. "Natural" human leukocyte antigen antibodies found in nonalloimmunized healthy males. Transplantation 2008;86:1111-5. [Crossref] [PubMed]
- Susal C, Roelen DL, Fischer G, et al. Algorithms for the determination of unacceptable HLA antigen mismatches in kidney transplant recipients. Tissue Antigens 2013;82:83-92. [Crossref] [PubMed]
- Fuggle SV, Martin S. Tools for human leukocyte antigen antibody detection and their application to transplanting sensitized patients. Transplantation 2008;86:384-90. [Crossref] [PubMed]
- Bostock IC, Alberu J, Arvizu A, et al. Probability of deceased donor kidney transplantation based on % PRA. Transpl Immunol 2013;28:154-8. [Crossref] [PubMed]
- Snyder LD, Gray AL, Reynolds JM, et al. Antibody desensitization therapy in highly sensitized lung transplant candidates. Am J Transplant 2014;14:849-56. [Crossref] [PubMed]
- Locke JE, Zachary AA, Warren DS, et al. Proinflammatory events are associated with significant increases in breadth and strength of HLA-specific antibody. Am J Transplant 2009;9:2136-9. [Crossref] [PubMed]
- Augustine JJ, Woodside KJ, Padiyar A, et al. Independent of nephrectomy, weaning immunosuppression leads to late sensitization after kidney transplant failure. Transplantation 2012;94:738-43. [Crossref] [PubMed]
- Duquesnoy RJ. A structurally based approach to determine HLA compatibility at the humoral immune level. Hum Immunol 2006;67:847-62. [Crossref] [PubMed]
- Sinha N, Mohan S, Lipschultz CA, et al. Differences in electrostatic properties at antibody-antigen binding sites: implications for specificity and cross-reactivity. Biophys J 2002;83:2946-68. [Crossref] [PubMed]
- Kosmoliaptsis V, Chaudhry AN, Sharples LD, et al. Predicting HLA class I alloantigen immunogenicity from the number and physiochemical properties of amino acid polymorphisms. Transplantation 2009;88:791-8. [Crossref] [PubMed]
- Doyle HR, Marino IR, Morelli F, et al. Assessing risk in liver transplantation. Special reference to the significance of a positive cytotoxic crossmatch. Ann Surg 1996;224:168-77. [Crossref] [PubMed]
- Smith JD, Ibrahim MW, Newell H, et al. Pre-transplant donor HLA-specific antibodies: characteristics causing detrimental effects on survival after lung transplantation. J Heart Lung Transplant 2014;33:1074-82. [Crossref] [PubMed]
- Loupy A, Lefaucheur C, Vernerey D, et al. Complement-binding anti-HLA antibodies and kidney-allograft survival. N Engl J Med 2013;369:1215-26. [Crossref] [PubMed]
- Sicard A, Ducreux S, Rabeyrin M, et al. Detection of C3d-binding donor-specific anti-HLA antibodies at diagnosis of humoral rejection predicts renal graft loss. J Am Soc Nephrol 2015;26:457-67. [Crossref] [PubMed]
- Lowe D, Higgins R, Zehnder D, et al. Significant IgG subclass heterogeneity in HLA-specific antibodies: Implications for pathogenicity, prognosis, and the rejection response. Hum Immunol 2013;74:666-72. [Crossref] [PubMed]
- Lefaucheur C, Viglietti D, Bentlejewski C, et al. IgG Donor-Specific Anti-Human HLA Antibody Subclasses and Kidney Allograft Antibody-Mediated Injury. J Am Soc Nephrol 2016;27:293-304. [Crossref] [PubMed]
- Montgomery RA, Warren DS, Segev DL, et al. HLA incompatible renal transplantation. Curr Opin Organ Transplant 2012;17:386-92. [Crossref] [PubMed]
- Biglarnia AR, Huber-Lang M, Mohlin C, et al. The multifaceted role of complement in kidney transplantation. Nat Rev Nephrol 2018;14:767-81. [Crossref] [PubMed]
- Abu Jawdeh BG, Cuffy MC, Alloway RR, et al. Desensitization in kidney transplantation: review and future perspectives. Clin Transplant 2014;28:494-507. [Crossref] [PubMed]
- Malvezzi P, Jouve T, Noble J, et al. Desensitization in the Setting of HLA-Incompatible Kidney Transplant. Exp Clin Transplant 2018;16:367-75. [PubMed]
- Cole RM, Kobashigawa JA. Desensitization Strategies Pre- and Post-Cardiac Transplantation. Curr Treat Options Cardiovasc Med 2016;18:8. [Crossref] [PubMed]
- Reinsmoen NL, Patel J, Mirocha J, et al. Optimizing transplantation of sensitized heart candidates using 4 antibody detection assays to prioritize the assignment of unacceptable antigens. J Heart Lung Transplant 2016;35:165-72. [Crossref] [PubMed]
- Geft D, Kobashigawa J. Current concepts for sensitized patients before transplantation. Curr Opin Organ Transplant 2017;22:236-41. [Crossref] [PubMed]
- Kim M, Martin ST, Townsend KR, et al. Antibody-mediated rejection in kidney transplantation: a review of pathophysiology, diagnosis, and treatment options. Pharmacotherapy 2014;34:733-44. [Crossref] [PubMed]
- Tambur AR, Campbell P, Claas FH, et al. Sensitization in Transplantation: Assessment of Risk (STAR) 2017 Working Group Meeting Report. Am J Transplant 2018;18:1604-14. [Crossref] [PubMed]


