Gut microbiome beats two to zero host genome
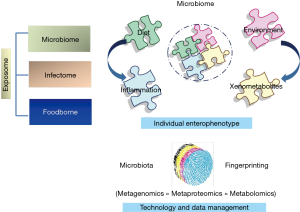
The gut microbiota and its genomic scaffold, exceeding the human one nearly 500 times, substantially affect human health and diseases. Host-microbe interactions, exerted through microbial biochemical and immunological activities, contain pathogen burden, control neurological and endocrine signalling, enterocyte wellness, energy biosynthesis, gut dysbiosis, complement gaps of host metabolic pathways, finally contributing to human physiology and disease within the gastrointestinal district and through gut-liver and gut-brain axis (1). Regardless substantial advances in correlating microbiota modulation and perturbation with various influencers, the specific impact of internal and external stimuli need to be definitely assigned though causality relationships beyond correlative measures. Host origin or genetic background, age, sanitation, delivery mode, breast-feeding and weaning, infections, diet, drugs, but also exercise, sleep, stress have been reviewed in a wide range of recent literature (2). All external (i.e., food, pathogens) and internal (i.e., microbiota) host modulating factors, both, can be conceptually synthetized by the term “exposome” which we are exposed to in the lifetime and which drives both individual enterophenotypes and disease phenotypes (3). Integrative descriptive and functional charts of microbiota allow the description of the microbiota-host Holobionts system, relationship that can be employed in understanding personalized physiology and nutrition, thus providing patient-tailored therapies (Figure 1). To investigate microbiota unbalance and passage from healthy to disease or unhealthy status, individual baseline compositions and variations within an individual's own range are required.
However, to establish dysbiosis traits at population level, the selection of large individual cohorts from different origins is mandatory, defining baselines of microbiota profiles under steady or eubiotic states. Big healthy population reservoirs, with subjects stratified with reference to age, diet, drugs and anthropometric measurements can constitute reference populations. From these groups of healthy subjects, it is possible to assess microbial dynamics, ecological networks and interspecies interactions in a way to have ecological signatures of the dynamic and thoroughly individualized ecosystems (4). Despite elevated levels of inter-individual variegation in either species cataloguing and quantification, other parameters such as bacterial growth rates and intra- and inter-species reciprocal relationship in microbial ecosystems of distinct individuals may reveal general dynamics, with the inter-individual variation mainly generated from dissimilarity in the sets of colonizing species (5). In the study by Bashan et al., 2016 (4), authors developed a computing procedure to describe human gut microbial dynamics to cross-sectional evidence from two large-scale metagenomic studies (6) that assessed universal dynamics for gut microbiota. Universality of gut microbiota dynamics/network definitely assist in the comprehension of the mechanisms shaping human microbial ecosystems, and, hence, allow to outlining microbiome-based therapies (7). Once general microbiota architecture rules will be definitely highlighted for healthy populations groups, the extension to the definition of healthy population microbiota profiling and metagenomics-based personalized gut microbiome variants will become affordable (8). These kind of studies on gut microbiota may prepare the way to the identification of disease risk factors, microbiota-linked disease biomarkers and disease prediction models.
The other central issue is how microbiome structure is set by host genetics. Former studies have highlighted numerous heritable microbial taxa (9), but without investigate quantification of specific bacterial abundances. Furthermore, other studies have identified associations between host single nucleotide polymorphisms (SNPs) and single bacterial taxa or pathways (10-12). Nevertheless, the major number of previous associations resulted not statistically significant following filtering by multiple regression test correction (13).
The study by Rothschild et al., appeared in a recent issue of Nature (14), has evaluated microbial-genetic associations on a cohort of 1,046 healthy Israeli adults by exploiting targeted-metagenomics of gut microbiomes, genotypes, anthropometric and blood laboratory values, and nutritional habits. The results were duplicated on the “validation” subject group LifeLines DEEP (LLD) cohort, consisting of 836 Dutch individuals (11). However, because twin investigations are perfect for heritability evaluation (15), authors also analysed datasets from a 2,252 Twins UK cohort (10). By exploiting correlations between genetic ancestry, SNPs and microbiome composition, the lack of relationship between microbiome profile and genetic background was demonstrated.
However, a “microbiome-association” scale, designed by analogy with genetic heritability, was introduced to quantify the overall microbiome-host phenotype association (1) after accounting for host genetics. These associations, regardless being correlative measures and not causality indexes, assigned the fraction of microbiome diversity variance to a whole 22–36% range, accounting for the 25% to the body mass index and glycaemic status; for the 22% to fasting glucose levels; for the 36% to levels of high-density lipoprotein (HDL) cholesterol and lactose consumption; for the 29% to waist circumference; for the 27% to hip circumference; and for the 24% to waist-hip ratio.
Furthermore, also microbiome-environment association computations were performed on past or present household sharing, which was described as partly determining gut microbiome profiling, while only few data for microbiome patterns similarity within relatives with no previous domestic sharing was inferred. The results were consistent with previous studies, including a recent study on twin showing that microbiome chronologically evolve more genetically divergent when living apart (12).
In summary, the genotype and microbiome data from the 1,046 healthy subjects’ cohort, characterized by with numerous different ancestral origins and a rather shared environment, allowed the group of Rothschild et al., to conclude that gut microbiome is not significantly linked to genetic ancestry, and that host genetics plays a slight role in shaping microbiome profile. Significant resemblances in microbiomes patterns of genetically not related subjects who share a household were showed, and above 20% of the inter-person microbiome variability was related with factors such as diet, drugs and anthropometric values.
In conclusion, the study by Rothschild et al., indicates that microbiome data can extensively get better the prediction for many disease-related human traits, such as glucose and obesity values, compared to models that use only host genetic. Definitively, the gut microbiome beats two to zero host genome to interpret host phenotype and the exposome definitely dominates over host genetics in modulating gut microbiota
All together, these evidences concur in considering microbiome-based therapies valuable tools in clinical outcomes across diverse genetic backgrounds.
Acknowledgments
None.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Lynch SV, Pedersen O. The Human Intestinal Microbiome in Health and Disease. N Engl J Med 2016;375:2369-79. [Crossref] [PubMed]
- Bäckhed F, Roswall J, Peng Y, et al. Dynamics and Stabilization of the Human Gut Microbiome during the First Year of Life. Cell Host Microbe 2015;17:852. [Crossref] [PubMed]
- Putignani L, Dallapiccola B. Foodomics as part of the host-microbiota-exposome interplay. J Proteomics 2016;147:3-20. [Crossref] [PubMed]
- Bashan A, Gibson TE, Friedman J, et al. Universality of human microbial dynamics. Nature 2016;534:259-62. [Crossref] [PubMed]
- Costello EK, Stagaman K, Dethlefsen L, et al. The application of ecological theory toward an understanding of the human microbiome. Science 2012;336:1255-62. [Crossref] [PubMed]
- Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 2012;486:207-14. [Crossref] [PubMed]
- Lemon KP, Armitage GC, Relman DA, et al. Microbiota-targeted therapies: an ecological perspective. Sci Transl Med 2012;4:137rv5. [Crossref] [PubMed]
- Schloissnig S, Arumugam M, Sunagawa S, et al. Genomic variation landscape of the human gut microbiome. Nature 2013;493:45-50. [Crossref] [PubMed]
- Turpin W, Espin-Garcia O, Xu W, et al. Association of host genome with intestinal microbial composition in a large healthy cohort. Nat Genet 2016;48:1413-7. [Crossref] [PubMed]
- Goodrich JK, Davenport ER, Beaumont M, et al. Genetic Determinants of the Gut Microbiome in UK Twins. Cell Host Microbe 2016;19:731-43. [Crossref] [PubMed]
- Bonder MJ, Kurilshikov A, Tigchelaar EF, et al. The effect of host genetics on the gut microbiome. Nat Genet 2016;48:1407-12. [Crossref] [PubMed]
- Xie H, Guo R, Zhong H, et al. Shotgun Metagenomics of 250 Adult Twins Reveals Genetic and Environmental Impacts on the Gut Microbiome. Cell Syst 2016;3:572-84.e3. [Crossref] [PubMed]
- Kurilshikov A, Wijmenga C, Fu J, et al. Host Genetics and Gut Microbiome: Challenges and Perspectives. Trends Immunol 2017;38:633-47. [Crossref] [PubMed]
- Rothschild D, Weissbrod O, Barkan E, et al. Environment dominates over host genetics in shaping human gut microbiota. Nature 2018;555:210-5. [Crossref] [PubMed]
- Visscher PM, Goddard ME. A general unified framework to assess the sampling variance of heritability estimates using pedigree or marker-based relationships. Genetics 2015;199:223-32. [Crossref] [PubMed]