Comprehensive molecular profiling of extrahepatic cholangiocarcinoma in Chinese population and potential targets for clinical practice
Introduction
Cholangiocarcinoma (CCA) is a heterogeneous malignancy of the biliary tract epithelium with an increasing incidence and mortality (1,2). CCA arising from extrahepatic biliary system was as extrahepatic cholangiocarcinoma (ECC) (3) which accounts for about 90% of the CCA cases (4,5). The risk factors for ECC include primary sclerosing cholangitis, ulcerative colitis, abnormal choledochopancreatic junction and liver flukes (6).
Complete resection with negative margins is the only potentially curative treatment for resectable ECC but most of CCA patients were in the advanced-stage at presentation (5,7). Besides, the 5-year overall survival of patients was only 27% if R0 resection was achieved (4). Based on ABC-02 (8) and other trials (9,10), the combination of gemcitabine and cisplatin is considered to be the standard of care for first-line chemotherapy for patients with advanced or metastatic biliary tract cancers. But the median survival of this combination was only 11.7 months for patients with locally advanced or metastatic CCA, gallbladder cancer or ampullary cancer (8). Recently, molecular targeted therapies have been developed for the CCA treatment based on the genetic information. Small molecule inhibitors of FGFR such as BGJ398, erdafitinib and derazantinib are being investigated in clinical trials (11,12). The other molecular targets for CCA encompass IDH1, HER2 and CDKN2A (13-15). The molecular understanding of the genetic alterations of CCA holds promise in advancing the field of personalized therapeutic intervention.
Several studies focused on the molecular profiling of CCA in the intra- and extrahepatic anatomical sites to identify recurrent driver gene alterations for systemic therapy. Previous study revealed the distinct genomic profiling of ECC which is significantly different from intrahepatic CCA. The recurrent mutations of IDH1/2 and FGFR2 rearrangements/fusions in the intrahepatic CCA were detected with low frequency or even not detected in the ECC patients (16). Lowery et al. demonstrated that KRAS, SMAD4, TP53 and STK11 were significantly mutated in the ECC relative to the intrahepatic CCA (17). But little evidence revealed the genomic characteristics for ECC in Chinese population. It has been reported that patients from different ethnicity showed the distinct genomic profiling in other carcinomas (18).
Next generation sequencing is an ideal tool for the targeted therapy in the era of personalized medicine (19) In this study, next generation sequencing was performed for 80 Chinese ECC patients to elucidate the genomic characteristics which may benefit further precise treatment.
Methods
Patients
Patients were enrolled from multiple accredited clinical hospitals between 2015 and 2018. Each patient had a confirmed histologic diagnosis of ECC. Results from 80 Chinese patients with ECC were available at the time of analysis. Clinical data were recorded, including age, gender, family and personal history of malignancy and treatments. This study was approved by the institutional review board at the local sites. Informed written consent was obtained from each patient.
Sample preparation
Patients’ tissue samples were formalin fixed, paraffin-embedded (FFPE) in accredited clinical hospitals. The histologic sections were retrieved. Independent pathologists reviewed all the tumor samples. The percentage of tumor cells for each sample was ensured to be more than 20%. And then 50–250 ng DNA was extracted from the unstained tissue sections for subsequent analysis of genetic alterations.
Genetic analysis
Comprehensive profiling of genomic alterations was detected by next generation sequencing. Hybridization capture of 7,029 exons of 450 cancer-related genes and selected introns of 35 genes commonly rearranged in cancers were applied to the extracted DNA. For subsequent sequencing, a minimum of 40 ng DNA was required. APA Hyper Prep Kit (KAPA Biosystems Inc., Basel, Switzerland) was used in the process. Sequencing of the captured libraries was run on an Illumina NextSeq 500. For estimation of sequencing error rate, a PhiX spike-in was added as an external control, and a percentage of mismatches less than 4% was qualified.
In order to ensure the overall quality of the study, post-sequencing quality control metrics including contamination ratio, total usable sequences, low quality reads ratio, ratio of aligned bases on target reads, and mean sequencing depth were monitored. Samples and/or sequencing run that failed to meet any one of requirement above were excluded. Analysis of genomic DNA from tumor tissue and matched blood control were performed according to previous reports (20-22). Genomic alterations including single-nucleotide variations (SNV), short and long insertions and deletions (indels), copy number alternations, and gene rearrangements/fusions were identified.
Microsatellite instability (MSI) status was identified in all cases. According to the MSI score, samples were classified into three groups, including MSI-high with ≥2 instable microsatellite loci, MSI-low with only 1 instable loci and microsatellite stable (MSS) with no loci instability. If identification for a sample is not clear, analysis was repeated to clarify the status. The MSI-high result was confirmed further by polymerase chain reaction validation.
The tumor mutational burden (TMB) was estimated by analyzing somatic mutations including coding SNVs and indels per mega-base of the panel sequences examined. Driver gene mutations and known germline alterations in the single nucleotide polymorphism database (dbSNP) were excluded, following the method of Chalmers et al. (23).
Statistics analysis
All statistics analysis was performed on R software. For evaluating the association of clinical characteristics or genes, Fisher’s exact tests were performed. False discovery rate (FDR) corrected P value (Q value) was also calculated. For all the analysis, only a P value below 0.05 was considered significant.
Results
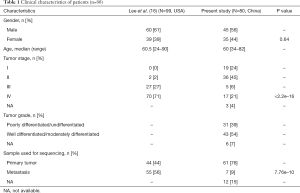
Samples from 80 individual patients with ECC were analyzed and the clinical characteristics of patients were shown in Table 1. The samples were obtained from 45 males and 35 females with a median age of 60 years (range, 34–82). Nineteen (24%) ECC patients were at stage I, 36 (45%) stage II, 5 (6%) stage III and 17 (21%) stage IV tumors. There were 31 (39%) patients with poorly differentiated/undifferentiated tumors and 43 (54%) patients with well/moderately differentiated tumors. Majority of tumor samples (61, 76%) were obtained from the primary bile duct tumor.
Full table
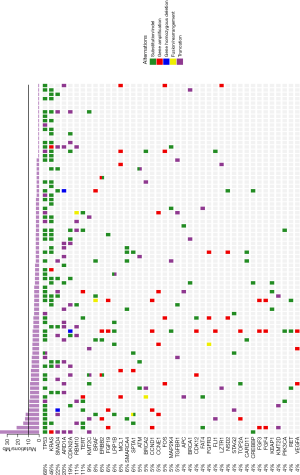
A total of 471 genetic alterations including both somatic and germline mutations were identified among 80 samples with an average of six mutations per sample in 177 genes. The molecular profiling and TMB results were displayed in Figure 1. The most commonly mutated genes were TP53 (68%), KRAS (46%), SMAD4 (22%), ARID1A (20%), CDKN2A (19%), RBM10 (11%), TERT (11%) and KMT2C (9%). Only one patient harbored none somatic mutation. Two patients harbored IDH1/2 mutations. Nineteen structural alterations were identified in eight patients (10%). One patient had NXPH1-BRAF rearrangements/fusion events indicating activation of BRAF. High TMB (>10 muts/Mb) was only observed in six patients. All patients had signature of microsatellite stability (see details in the Methods).
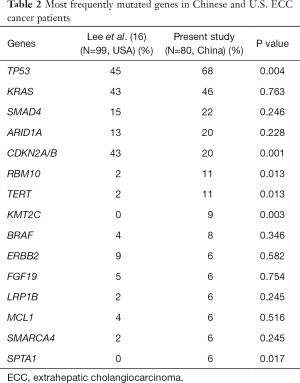
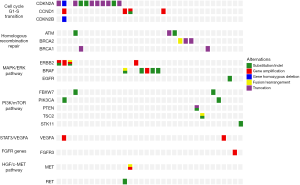
A comparative analysis was conducted between our results and the other results consisting of USA population (16). Distinct patterns of genetic alterations were investigated (Table 2). TP53, RBM10, TERT, KMT2C and SPTA1 alterations were more commonly seen in our study while CDKN2A was more commonly seen in USA population (P value <0.05). We also observed mutual exclusivity between commonly altered genes including ARID1A:TP53, KRAS:LRP1B and NF2:TP53 (Table 3). Potentially actionable genetic alterations: 43 patients (43%) had at least one actionable alteration which targeted inhibitors have demonstrated compelling clinical activity in CCA or other cancer types. Actionable genetic alterations were based on databases such as oncokb (www.oncokb.org), cosmic (cancer.sanger.ac.uk/cosmic), clinicaltrials (cancer.sanger.ac.uk/cosmic/) and pubmed (www.ncbi.nlm.nih.gov/pubmed). The most frequently recurrent actionable alterations included CDKN2A (n=11), BRAF (n=5), ERBB2 (n=4), BRCA1/2 (n=5), CCND1 (n=4) and ATM (n=3). Several patients had more than one potentially actionable genetic alteration: three patients with more than 3 (including 3) actionable alterations and three patients with 2 actionable alterations. Additional potentially actionable alterations presented at low frequency, including PIK3CA (n=2), PTEN (n=1), and MET (n=1). Potentially actionable alterations were enriched in the G1-S transition, homologous recombination repair, MAPK/ERK pathway, STAT3/VEGFA, PI3K/mTOR pathway and HGF/c-MET pathway (Figure 2).
Full table

Full table
Discussion
In this study, DNA targeted sequencing of cancer associated genes was performed for 80 ECC patients. FFPE tissue obtained from core needle biopsy or resected specimen of primary or metastatic sites of disease was used for genomic profiling. In almost 43% of patients with ECC, at least one actionable genetic alteration was identified. To our knowledge, this is the first report about the molecular profiling of ECC using DNA targeted sequencing in Chinese population.
The mutation frequencies of top five genes in this study include TP53, KRAS, SMAD4, ARID1A and CDKN2A. The most common actionable alterations were loss of function of CDKN2A, activation of BRAF and amplification of ERBB2. Loss-of-function mutations of CDKN2A could lead to the activation of CDK4/6 (24). Several clinical trials have been performed for evaluating the activity of CDK4/6 inhibitor, palbociclib, in patients with lung cancer, urothelial cancer and breast cancer (25-27). However, no clinical trial of palbociclib therapy has been or is being performed for CCA especially for ECC. Palbociclib in combination with radiation therapy exerts anti-cancer effect on human hepatocellular carcinoma and CCA cell lines (28). Pan-mTOR inhibitor MLN0128 synergize with palbociclib to impair intrahepatic CCA growth in vitro and in vivo (29). Other CDK4/6 inhibitors, abemaciclib and ribociclib, were assessed in lung cancer and breast cancer (30,31).
BRAF mutations detected in our cohorts account for 7% of all ECC patients. But V600E mutation of BRAF was not observed. A phase II study was evaluated the BRAF inhibitor, trametinib, in patients with refractory or advanced biliary or gallbladder cancer or that was unresectable (NCT02042443). A phase II trial of BRAF inhibitor regorafenib as a single agent was evaluated in patients with chemotherapy-refractory, advanced, and metastatic biliary tract adenocarcinoma. Five of 43 patients treated with regorafenib achieved partial response and 19 patients had stable disease (32). Several phase II trials of BRAF inhibitor, sorafenib, alone or combination with other agents have been done for unselected CCA but had low activity or did not improve efficacy (33-35). A phase II trial of the anti-Her2 antibody–drug conjugate trastuzumab emtansine in advanced-stage CCA is currently ongoing (NCT02999672). Preliminary results from phase I/II studies of agents targeting CDKN2A, BRAF and ERBB2 mutations indicate that these agents have activity in molecularly selected populations.
In our cohort, all patients showed stable microsatellite, which was consistent with previous report (17). Our unselected patient population with ECC may be more reflective of the patients in need of novel systemic therapies. However, our data suggest that MSI-High may be a less common occurrence in patients with ECC than other carcinomas such as colorectal cancer. Fortunately, we observed seven patients with high TMB (TMB >10 muts/Mb). Given the recent FDA-approval of immune checkpoint blockade for TMB-H non-small lung cancer patients, TMB might be an indicative marker for these ECC patients.
Conclusions
In summary, we identified distinct genomic mutation profiles in Chinese ECC patients which indicate multiple potentially actionable genetic alterations. Nearly half of patients were identified with potentially actionable alterations, which suggest that molecular profiling could be applied in clinical practice to change the management of this fatal cancer in the future. Molecular profiling of patients with ECC should be considered for all patients to screen potential clinical trial candidates. For the rarity and heterogeneity of this disease, cooperation of academic centers and industry will accelerate the application of targeted- and immune-therapy in ECC.
Acknowledgments
None.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was approved by the institutional review board at the local sites (No. QDFYKYLLL-20161212). Informed written consent was obtained from each patient.
References
- Khan SA, Taylor-Robinson SD, Toledano MB, et al. Changing international trends in mortality rates for liver, biliary and pancreatic tumours. J Hepatol 2002;37:806-13. [Crossref] [PubMed]
- Saha SK, Zhu AX, Fuchs CS, et al. Forty-year trends in cholangiocarcinoma incidence in the U.S.: intrahepatic disease on the rise. Oncologist 2016;21:594-9. [Crossref] [PubMed]
- Hamilton SR, Aaltonen LA. WHO classification of tumours. Pathology and genetics of tumours of the digestive system. Lyon: IARC Press, 2000.
- DeOliveira ML, Cunningham SC, Cameron JL, et al. Cholangiocarcinoma: thirty-one-year experience with 564 patients at a single institution. Ann Surg 2007;245:755-62. [Crossref] [PubMed]
- Razumilava N, Gores GJ. Cholangiocarcinoma. Lancet 2014;383:2168-79. [Crossref] [PubMed]
- Bosman FT, Carneiro F, Hruban RH, et al. WHO classification of tumours of the digestive system. 4th ed. Geneva: World Health Organization, 2010:417.
- Jarnagin WR, Fong Y, DeMatteo RP, et al. Staging, resectability, and outcome in 225 patients with hilar cholangiocarcinoma. Ann Surg 2001;234:507-17; discussion 517-9. [Crossref] [PubMed]
- Valle J, Wasan H, Palmer DH, et al. Cisplatin plus gemcitabine versus gemcitabine for biliary tract cancer. N Engl J Med 2010;362:1273-81. [Crossref] [PubMed]
- Okusaka T, Nakachi K, Fukutomi A, et al. Gemcitabine alone or in combination with cisplatin in patients with biliary tract cancer: a comparative multicentre study in Japan. Br J Cancer 2010;103:469-74. [Crossref] [PubMed]
- Grenader T, Nash S, Plotkin Y, et al. Derived neutrophil lymphocyte ratio may predict benefit from cisplatin in the advanced biliary cancer: the ABC-02 and BT-22 studies. Ann Oncol 2015;26:1910-6. [Crossref] [PubMed]
- Javle MM, Shroff RT, Zhu A, et al. A phase 2 study of BGJ398 in patients (pts) with advanced or metastatic FGFR-altered cholangiocarcinoma (CCA) who failed or are intolerant to platinum-based chemotherapy. J Clin Oncol 2016;34:335. [Crossref]
- Tabernero J, Bahleda R, Dienstmann R, et al. Phase I dose-escalation study of JNJ-42756493, an oral pan-fibroblast growth factor receptor inhibitor, in patients with advanced solid tumors. J Clin Oncol 2015;33:3401-8. [Crossref] [PubMed]
- Burris H, Mellinghoff I, Maher E, et al. Abstract PL04-05: The first reported results of AG-120, a first-in-class, potent inhibitor of the IDH1 mutant protein, in a Phase I study of patients with advanced IDH1-mutant solid tumors, including gliomas. Molecular Cancer Therapeutics 2015;14:PL04-5.
- Javle M, Churi C, Kang HC, et al. HER2/neu-directed therapy for biliary tract cancer. J Hematol Oncol 2015;8:58. [Crossref] [PubMed]
- Ross JS, Wang K, Gay L, et al. New routes to targeted therapy of intrahepatic cholangiocarcinomas revealed by next-generation sequencing. Oncologist 2014;19:235-42. [Crossref] [PubMed]
- Lee H, Wang K, Johnson A, et al. Comprehensive genomic profiling of extrahepatic cholangiocarcinoma reveals a long tail of therapeutic targets. J Clin Pathol 2016;69:403-8. [Crossref] [PubMed]
- Lowery MA, Ptashkin R, Jordan E, et al. Comprehensive molecular profiling of intrahepatic and extrahepatic cholangiocarcinomas: potential targets for intervention. Clin Cancer Res 2018;24:4154-61. [Crossref] [PubMed]
- Sun Y, Ren Y, Fang Z, et al. Lung adenocarcinoma from East Asian never-smokers is a disease largely defined by targetable oncogenic mutant kinases. J Clin Oncol 2010;28:4616-20. [Crossref] [PubMed]
- Friedman AA, Letai A, Fisher DE, et al. Precision medicine for cancer with next-generation functional diagnostics. Nat Rev Cancer 2015;15:747-56. [Crossref] [PubMed]
- Frampton GM, Fichtenholtz A, Otto GA, et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol 2013;31:1023-31. [Crossref] [PubMed]
- Ong CK, Subimerb C, Pairojkul C, et al. Exome sequencing of liver fluke-associated cholangiocarcinoma. Nat Genet 2012;44:690-3. [Crossref] [PubMed]
- Nakamura H, Arai Y, Totoki Y, et al. Genomic spectra of biliary tract cancer. Nat Genet 2015;47:1003-10. [Crossref] [PubMed]
- Chalmers ZR, Connelly CF, Fabrizio D, et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med 2017;9:34. [Crossref] [PubMed]
- VanArsdale T, Boshoff C, Arndt KT, et al. Molecular pathways: targeting the cyclin D-CDK4/6 axis for cancer treatment. Clin Cancer Res 2015;21:2905-10. [Crossref] [PubMed]
- Zhou J, Zhang S, Chen X, et al. Palbociclib, a selective CDK4/6 inhibitor, enhances the effect of selumetinib in RAS-driven non-small cell lung cancer. Cancer Lett 2017;408:130-7. [Crossref] [PubMed]
- Rose TL, Chism DD, Alva AS, et al. Phase II trial of palbociclib in patients with metastatic urothelial cancer after failure of first-line chemotherapy. Br J Cancer 2018;119:801-7. [Crossref] [PubMed]
- Finn RS, Crown JP, Lang I, et al. The cyclin-dependent kinase 4/6 inhibitor palbociclib in combination with letrozole versus letrozole alone as first-line treatment of oestrogen receptor-positive, HER2-negative, advanced breast cancer (PALOMA-1/TRIO-18): a randomised phase 2 study. Lancet Oncol 2015;16:25-35. [Crossref] [PubMed]
- Huang CY, Hsieh FS, Wang CY, et al. Palbociclib enhances radiosensitivity of hepatocellular carcinoma and cholangiocarcinoma via inhibiting ataxia telangiectasia-mutated kinase-mediated DNA damage response. Eur J Cancer 2018;102:10-22. [Crossref] [PubMed]
- Song X, Liu X, Wang H, et al. Combined CDK4/6 and Pan-mTOR inhibition is synergistic against intrahepatic cholangiocarcinoma. Clin Cancer Res 2019;25:403-13. [Crossref] [PubMed]
- Kim ES, Kelly K, Paz-Ares LG, et al. Abemaciclib in combination with single-agent options in patients with stage IV non-small cell lung cancer: a phase Ib study. Clin Cancer Res 2018;24:5543-51. [Crossref] [PubMed]
- Hortobagyi GN, Stemmer SM, Burris HA, et al. Updated results from MONALEESA-2, a phase III trial of first-line ribociclib plus letrozole versus placebo plus letrozole in hormone receptor-positive, HER2-negative advanced breast cancer. Ann Oncol 2018;29:1541-7. [PubMed]
- Sun W, Patel A, Normolle D, et al. A phase 2 trial of regorafenib as a single agent in patients with chemotherapy-refractory, advanced, and metastatic biliary tract adenocarcinoma. Cancer 2019;125:902-9. [Crossref] [PubMed]
- Lee JK, Capanu M, O'Reilly EM, et al. A phase II study of gemcitabine and cisplatin plus sorafenib in patients with advanced biliary adenocarcinomas. Br J Cancer 2013;109:915-9. [Crossref] [PubMed]
- El-Khoueiry AB, Rankin C, Siegel AB, et al. S0941: a phase 2 SWOG study of sorafenib and erlotinib in patients with advanced gallbladder carcinoma or cholangiocarcinoma. Br J Cancer 2014;110:882-7. [Crossref] [PubMed]
- Bengala C, Bertolini F, Malavasi N, et al. Sorafenib in patients with advanced biliary tract carcinoma: a phase II trial. Br J Cancer 2010;102:68-72. [Crossref] [PubMed]