Decoding multifocal hepatocellular carcinoma: an opportune pursuit
Hepatocellular carcinoma (HCC) is the fifth most common cancer in men, accounting for more than 500,000 deaths per year worldwide (1). Approximately 70% to 90% of all cases of HCC occur in cirrhosis due to chronic infection by hepatitis B virus (HBV) and hepatitis C virus (HCV), toxic injury from excessive alcohol consumption, or metabolic liver disease primarily associated with obesity and diabetes (2). Long-term prognosis of HCC remains dire with 5-year survival rates hovering around 12% for all stages combined, although treatment interventions applied at early stages of HCC provide dramatically better results and justify regular surveillance and aggressive therapy (3). Accordingly, HCC staging has been linked to specific treatment strategies such as liver transplantation, surgical resection, and locoregional therapies in order to optimize clinical outcomes (4).
Liver resection is one of the most efficient interventions for the treatment of HCC, with 5-year survival rates ranging between 38% and 61% (5). Regrettably, about 75% of all HCC cases present as multiple intrahepatic tumors at the time of initial diagnosis, which may preclude surgical interventions with curative intent due to insufficient functional reserve of the remaining liver (6). Even if surgery can be safely performed, however, therapeutic success is limited by postoperative recurrence of HCC, which may reach 70% to 80% within 5 years. This is perhaps not surprising since cirrhosis is associated with a high risk for developing HCC and the chronically diseased residual liver tissue continues to have a malignant potential.
Postoperative recurrences of HCC can be addressed by complex surgical strategies that include repeated hepatic resection or salvage liver transplantation, but success for these interventions remains variable and difficult to predict (7). A likely reason for this heterogeneity is that the pathogenesis of multiple HCC includes at least 2 very different mechanisms (Figure 1). Multicentrically occurring HCC (MO-HCC) represents a polyclonal process of de novo hepatocarcinogenesis with primary tumor foci that have a clonal origin independent from each other and the formerly resected malignancy (8). Postsurgical recurrence of MO-HCC may respond well to additional surgery or loco-regional therapy and these efforts are often limited only by the functional hepatic reserve (9). HCC with intrahepatic metastases (IM-HCC), on the other hand, originates from the dissemination of tumor cells with a common clonal origin and with increasingly aggressive biological behavior. IM-HCC seems to recur early following surgical resection and carries a grim prognosis despite heroic interventions (10).
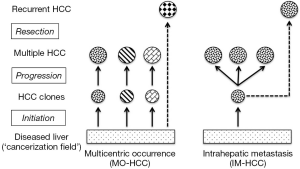
One of the key issues in the management of multiple HCC is therefore our ability to distinguish metastatic from multicentric hepatocarcinogenesis. This distinction may allow us to provide a more reliable prognosis and determine how far we should pursue therapeutic interventions with a curative intent (9). So far, differentiation of IM-HCC and MO-HCC (in the absence of extrahepatic spread, which would of course obviate this exercise) has been mostly based on histopathological findings as reported by the Liver Cancer Study Group of Japan (11,12). For instance, well-differentiated foci of recurrent HCC are more likely to originate from a de novo process (i.e., MO-HCC), while poor differentiation and invasive features point to metastatic dissemination (i.e., IM-HCC). Analysis of HBV-DNA integration into hepatocytes to determine the clonal origin of HCC may provide further clues about the mechanism of recurrence in cases associated with chronic hepatitis B (12). Furthermore, markers of tumor clonality may be obtained from frequently mutated proteins such as p53, selective X-chromosome inactivation pattern, loss of heterozygosity of microsatellite DNA loci, and chromosomal aberrations analyzed by comparative genomic hybridization (11,13). However, these diagnostic approaches have not yet yielded sufficient knowledge to become routine clinical practice and to guide the management of multiple HCC.
Novel molecular markers that reliably identify the pathomechanism of multiple HCC are therefore urgently needed. Fortunately, emerging biomedical technologies of systems biology have provided the impetus for achieving this objective. Next-generation sequencing and powerful computational tools increasingly allow the comprehensive characterization of cancers and link molecular and clinical phenotypes to better prognostication and optimized therapeutic interventions (14). In a recent issue of the Journal of Hepatology, Miao et al. have applied these principles to the management conundrum of multiple HCC (15). These authors utilized whole-genome and transcriptome sequencing to retrospectively identify biomarkers of tumor clonality and to associate the data with clinical outcomes in a cohort of Chinese patients with HBV-related HCC.
In their multi-omics analysis, Miao et al. initially selected two cases of multifocal hepatoma with disparate clinical courses following surgical resection. Patient I (PI) had multiple foci of poorly differentiated HCC in a cirrhotic liver and died 3 months after surgery. By contrast, patient II (PII) had multifocal, well-differentiated HCC in a non-cirrhotic liver and remained symptom-free at 2 years after surgery. Whole-genome sequencing revealed different patterns of HBV integration in these patients as all tumors in PI had a single HBV integration site, while two distinct tumor-initiating clones were identified in PII. Differences in clonality of PI and PII tissues were further confirmed by analysis of somatic mutation profiles and genomic structural variations that were validated by PCR and Sanger sequencing. Phylogenetic trees of PI tumors constructed from these data were similar to each other but far removed from the germ line, while the genetic variations found in PII tumors were best explained by the synchronous development of distinct clones. From these findings, Miao et al. concluded that PI and PII were prototype cases of IM-HCC and MO-HCC, respectively (15).
Miao et al. subsequently performed transcriptome analysis to characterize protein-coding gene expression in PI and PII tumors. As expected, similarities between the mRNA sets were more pronounced in PI than in PII tissue samples, further suggesting that multiple foci of HCC in PI were indeed the result of a monoclonal (i.e., metastatic) process. Subsequent functional enrichment mapping of differentially expressed genes of PI and PII indicated important topological differences in various gene function modules. According to this network analysis, essential changes in all PI tumors were comparable with upregulation of genes involved in cytoskeletal remodeling and extracellular matrix organization in PI satellite tumors, consistent with a metastatic signature. On the other hand, PII tumors displayed two distinct transcriptome patterns with some overlaps for tumorigenesis hallmarks such as negative regulation of apoptosis (15).
As a next step, Miao et al. utilized their transcriptome data to find genes with markedly different gene expression in PI and PII tumor tissues. Expression patterns of six genes with the most pronounced alterations (HAL, SFN, KIF15, TTK, BUB1, and MCM4) were analyzed against various clinico-pathological characteristics and postsurgical HCC recurrence in a cohort of 174 patients with HBV-related single or multifocal HCC. Interestingly, expression of TTK was found to have a strong reverse association with favorable postoperative prognosis in this cohort (15). The TTK gene encodes a dual-specificity kinase (also known as monopolar spindle 1 or Mps1 kinase) that phosphorylates serine, threonine, and tyrosine residues with a critical role in the regulation of cell division in normal and cancer cells (16). Mps1/TTK is required for normal chromosomal segregation and may serve a particular role in cancer by allowing sustained cell proliferation in the presence of aneuploidy (17). Consequently, Mps1/TTK is more than just a biomarker and has become a promising target in cancer therapy (18).
Miao et al. respectively linked the highly dissimilar clinical outcomes of HBV-related HCC in PI and PII to monoclonal and polyclonal cancer growth (15). Genomic integration of HBV increases the risk of hepatocarcinogenesis regardless of the presence of cirrhosis, and the authors reasonably assumed that HCC nodules in the non-cirrhotic liver of PII resulted from multiple occurrence, contrasted with a metastatic process in PI. However, multiple regenerative nodules in the remodeling cirrhotic liver may also serve as simultaneous sites of tumor initiation (19). This notion was corroborated by combined clinicopathological and genetic evaluation that distinguished IM-HCC vs. MO-HCC in 160 Chinese patients with HBV-related HCC and repeated surgical resection (9). Even though cirrhosis was more severe in the group of MO-HCC, patients in this earlier study had a significantly better disease-free survival, indicating that IM-HCC is not necessarily linked to the severity of underlying liver disease. Indeed, the combined effects of HBV integration and cirrhosis provide an intriguing example for the concept of ‘field cancerization’ in which multiple independent tumors may rise within a specific environment (20).
Could these observations provide new strategies with regards to the management of multiple HCC? If reliable biomarkers become available for the distinction of MO-HCC vs. IM-HCC, they may affect treatment algorithms for intermediate stage HCC with having multiple tumor foci in the liver. At the same time, there are issues that may limit enthusiasm for the surgical management of recurrent HCC. Repeated and generous resections may promote pro-oncogenic mechanisms associated with increased rates of liver regeneration whether or not cirrhosis has been established, accelerating further recurrence of HCC (12). Also, HCC may recur as a combination of multi-occurrence and intrahepatic metastasis, possibly calling for even more sophisticated biomarkers to guide clinical management.
Can we extrapolate the findings of Miao et al. to non-HBV-related HCC? There is evidence that the incidence of MO-HCC is significantly higher in HCV-positive patients compared to those with chronic HBV infection (9). These observations may reflect an accelerated rate of tumor progression due to concurrent oncogenic processes and a higher proportion of IM-HCC in HBV-associated cirrhosis. The findings that increased TTK mRNA levels predict a more aggressive course of HCC in patients with HBV-associated liver disease indicate that TTK may become a new biomarker for the presence of IM-HCC with a more aggressive clinical course, which includes a shorter interval of postoperative HCC recurrence. Importantly, Miao et al. also found a highly significant association between TTK gene expression and HBsAg-positivity (15). Future studies will determine whether upregulation of the mitotic checkpoint regulator TTK is a useful parameter in predicting the biological behavior of non-HBV-related HCC.
The quest to find biomarkers that reliably identify MO-HCC vs. IM-HCC is also about defining the biological characteristics of tumor initiation vs. progression. Clinical experience indicates that making this distinction has tremendous implications for the affected individuals. Miao et al. have taken a significant step towards applying the methods of multi-omics analysis and network medicine to track changes in the genome and transcriptome of liver cells linked to these two different aspects of tumorigenesis (15). Extrapolation of functional gene enrichment analysis has yielded a promising biomarker (TTK) to assist prognostication and guide the management of multifocal HCC. Our hope is that increasingly applying the tools of systems biology to the problem of HCC clonality will bring precision and efficacy beyond the current state of the art.
Acknowledgements
Disclosure: The author declares no conflict of interest.
References
- Jemal A, Bray F, Center MM, et al. Global cancer statistics. CA Cancer J Clin 2011;61:69-90. [PubMed]
- El-Serag HB. Hepatocellular carcinoma. N Engl J Med 2011;365:1118-27. [PubMed]
- Forner A, Reig ME, de Lope CR, et al. Current strategy for staging and treatment: the BCLC update and future prospects. Semin Liver Dis 2010;30:61-74. [PubMed]
- Pons F, Varela M, Llovet JM. Staging systems in hepatocellular carcinoma. HPB (Oxford) 2005;7:35-41. [PubMed]
- Morise Z, Kawabe N, Tomishige H, et al. Recent advances in the surgical treatment of hepatocellular carcinoma. World J Gastroenterol 2014;20:14381-92. [PubMed]
- Jelic S, Sotiropoulos GC; ESMO Guidelines Working Group. Hepatocellular carcinoma: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 2010;21 Suppl 5:v59-64. [PubMed]
- Fuks D, Dokmak S, Paradis V, et al. Benefit of initial resection of hepatocellular carcinoma followed by transplantation in case of recurrence: an intention-to-treat analysis. Hepatology 2012;55:132-40. [PubMed]
- Forner A, Gilabert M, Bruix J, et al. Treatment of intermediate-stage hepatocellular carcinoma. Nat Rev Clin Oncol 2014;11:525-35. [PubMed]
- Li Q, Wang J, Juzi JT, et al. Clonality analysis for multicentric origin and intrahepatic metastasis in recurrent and primary hepatocellular carcinoma. J Gastrointest Surg 2008;12:1540-7. [PubMed]
- Bruix J, Sherman M; American Association for the Study of Liver Diseases. Management of hepatocellular carcinoma: an update. Hepatology 2011;53:1020-2. [PubMed]
- Morimoto O, Nagano H, Sakon M, et al. Diagnosis of intrahepatic metastasis and multicentric carcinogenesis by microsatellite loss of heterozygosity in patients with multiple and recurrent hepatocellular carcinomas. J Hepatol 2003;39:215-21. [PubMed]
- Yamamoto T, Kajino K, Kudo M, et al. Determination of the clonal origin of multiple human hepatocellular carcinomas by cloning and polymerase chain reaction of the integrated hepatitis B virus DNA. Hepatology 1999;29:1446-52. [PubMed]
- Kawai S, Imazeki F, Yokosuka O, et al. Clonality in hepatocellular carcinoma: analysis of methylation pattern of polymorphic X-chromosome-linked phosphoglycerate kinase gene in females. Hepatology 1995;22:112-7. [PubMed]
- Fujimoto A, Totoki Y, Abe T, et al. Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nat Genet 2012;44:760-4. [PubMed]
- Miao R, Luo H, Zhou H, et al. Identification of prognostic biomarkers in hepatitis B virus-related hepatocellular carcinoma and stratification by integrative multi-omics analysis. J Hepatol 2014;61:840-9. [PubMed]
- Janssen A, van der Burg M, Szuhai K, et al. Chromosome segregation errors as a cause of DNA damage and structural chromosome aberrations. Science 2011;333:1895-8. [PubMed]
- Bursavich MG, Dastrup D, Shenderovich M, et al. Novel Mps1 kinase inhibitors: from purine to pyrrolopyrimidine and quinazoline leads. Bioorg Med Chem Lett 2013;23:6829-33. [PubMed]
- Aarts EO, Dogan K, Koehestanie P, et al. Long-term results after laparoscopic adjustable gastric banding: a mean fourteen year follow-up study. Surg Obes Relat Dis 2014;10:633-40. [PubMed]
- Piao Z, Park YN, Kim H, et al. Clonality of large regenerative nodules in liver cirrhosis. Liver 1997;17:251-6. [PubMed]
- Slaughter DP, Southwick HW, Smejkal W. Field cancerization in oral stratified squamous epithelium; clinical implications of multicentric origin. Cancer 1953;6:963-8. [PubMed]


