
Clinical and molecular heterogeneity associated with tumor sidedness in colorectal liver metastasis: a multicenter propensity cohort study
Highlight box
Key findings
• The prognostic value of tumor sidedness is restricted in patients with KRAS wild-type colorectal liver metastasis (CRLM).
• Right-sided CRLM more frequently harbors TP53, APC, KRAS, and BRAF alterations, and carry higher prevalence of pathogenic germline variants.
What is known and what is new?
• Colorectal cancer exhibits highly heterogeneity, with clinically and molecularly defined subgroups that differ in their prognosis.
• It remains unclear whether left-sided tumors are clinically and gnomically distinct from right-sided tumors in patients with CRLM.
What is the implication, and what should change now?
• Left-sided CRLM exhibits unique molecular profile distinct from right-sided CRLM that associates with their clinical heterogeneity, which emphasizes the importance of tumor sidedness-based stratification of CRLM, and have critical implications for designing therapeutic strategies.
Introduction
Colorectal cancer (CRC) metastasizes mainly to liver and its incidence is expected to rise further, which ranks as its most leading cause of cancer-related death worldwide (1-3). Tumor sidedness correlates with different biological and molecular features, with high heterogeneity defined subgroups that differ in their prognosis (4,5). The prognostic and predictive effects of primary tumor sidedness (PTS) in colorectal liver metastasis (CRLM) have been increasing recognized in recent years. However, the correlation between the prognostic impact of PTS and RAS mutational status is not fully elucidated, with discrepant results between studies.
Right-sidedness have been associated with resistance to anti-EGFR antibodies in RAS wild-type tumors. In the GALGB/SWOG 80405 trial comparing chemotherapy plus bevacizumab or cetuximab, KRAS wild-type patients with left-sidedness tumors experienced better outcomes with cetuximab than bevacizumab, while right-sidedness with KRAS wild-type tumors benefited more from bevacizumab than cetuximab (6). A recent meta-analysis of 66 relevant studies assembles a cohort of over 1 million patients with unresectable metastatic CRC (mCRC), and demonstrated that a left-sided primary tumor location was associated with a significantly reduced risk of death (7). A homogenous of pooled study included 9,277 mCRC patients from 12 first-line randomized trials in the ARCAD database confirmed that, among KRAS wild-type tumors, overall survival (OS) and disease-free survival (DFS) benefited from anti-EGFR were for left-sidedness, but not for right-sidedness (8). Thus, these suggest that the prognostic value of PTS in unresectable mCRC is restricted to the KRAS wild-type population.
However, the prognostic role of PTS may differ when it comes to resectable CRLM. Contrast to the analysis of unresectable mCRC, those in resectable CRLM remain largely unknown. There is currently no meta-analysis or clinical trials in patients with resectable CRLM that could conclude whether PTS is prognostic associated with RAS status.
Therefore, we aimed to investigate whether PTS is prognostic and explore the hypothesis that KRAS mutational status influences the prognostic value of PTS in resectable CRLM based on a large multicentric cohort study. Moreover, we performed whole exome sequencing (WES) to describe the somatic and germline landscapes between left-sidedness and right-sidedness. We present this article in accordance with the STROBE reporting checklist (available at https://hbsn.amegroups.com/article/view/10.21037/hbsn-22-285/rc).
Methods
Study population
We conducted a retrospective case-control study of 1,307 patients who underwent curative-intent resection of colorectal cancer and liver metastases between January 1, 2012 and December 31, 2020 from 3 independent centers in China, including Fudan University Shanghai Cancer Center, Shanghai (n=848), Fifth People’s Hospital of Shanghai Fudan University, Shanghai (n=242), and Shanghai Fengxian Central Hospital, Shanghai (n=217). This study confirmed to the standards of the Declaration of Helsinki (as revised in 2013). The study was approved by Independent Ethics Committee (IEC) of Fudan University Shanghai Cancer Center, Fifth People’s Hospital of Shanghai Fudan University, and Shanghai Fengxian Central Hospital. Written informed consent was obtained from the patients for the use of their clinical data and formalin-fixed paraffin-embedded tissue samples.
Standard demographic and clinicopathologic data were collected on each patient including sex, age, tumor characteristics, operative details, perioperative status, type and time of chemotherapy, molecular features, and date of last follow-up, and date of death. Primary tumor characteristics, including tumor location (left vs. right), American Joint Committee on Cancer T stage, nodal status, and tumor differentiation were recorded. Size, number, distribution of the hepatic metastases was also recorded. Tumor size and number were defined by the resection specimen. The largest lesion was used as the index lesion in the case of patients with multiple tumors.
The patients enrolled in the current study fulfilled the following criteria: (I) histologically confirmed CRLM, (II) receipt of complete resection and regular follow-up, (III) well-preserved liver function (Child-Pugh grad A or B), (IV) patients with both the left and right sided colorectal tumors, transverse colon primary tumors, or other malignant tumors were excluded from analysis, (V) patients without sidedness and molecular data were excluded from analysis. This resulted in a total of 1,307 patients included in the current analysis. In addition, a propensity score matching (PSM) with a 1:1 ratio matching was performed, and identified 481 pairs of matched patients formed the basis of this study (Figure 1A).
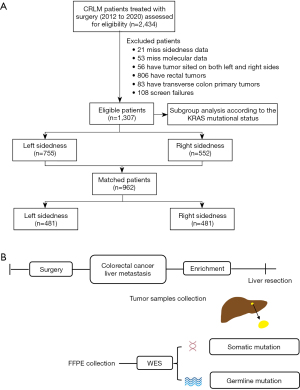
Comprehensive treatment strategy
Before surgery, all tumors were retrospectively assessed by expert groups who reviewed all pretreatment computed tomography scans or magnetic resonance imaging examinations to decide the treatment strategy. Patients with disease considered to be resectable were assigned to undergo liver resection with curative intent, with the aim of achieving complete resection while preserving as much normal, functional liver parenchyma as possible. It can take several forms advocated by current guidelines, such as (I) the classic approach, involving initial colorectal resection, interval chemotherapy, and liver resection or a liver-first approach with removal of the colorectal tumor as the final procedure, and (II) simultaneous removal of the liver and bowel tumors with or without neoadjuvant or adjuvant chemotherapy. According to the clinical risk score (CRS) proposed by Fong et al. (9), patients in the favorable prognosis group would be good candidates for upfront surgery, whereas for those in the poor prognosis group, expert recommendations are that chemotherapy should be administrated first.
Assessment of tumor sidedness
Information on tumor sidedness was obtained from the free-text surgery descriptions included in the case report forms and from the original pathology reports. Primary tumors originating in the splenic flexure, descending colon, or sigmoid colon were classified as left-sided CRLM. Primary tumors located in the cecum, ascending colon, or hepatic flexure were categorized as right-sided CRLM. We excluded patients with transverse colon primary tumors (with the exception of “splenic flexure” tumors, which were included in the left-sided group; the exception of “hepatic flexure” tumors, which were included in the right-sided group). First, it would allow for the comparison with the one of the largest studies to date on PTS in resectable CRLM, which employed this exclusion criterion. Second, it is often impossible to determine retrospectively from the pathologic report whether the primary tumor was located before or after the point that separates the first two-thirds from the final third of the transverse colon. Importantly, location compared to that point determines the embryologic origin (midgut vs. hindgut) of the tumor. Patients who had multiple primary tumors identified in both left- and right-sided locations were excluded.
KRAS mutation profiling
As previously described, the extracted DNA was evaluated for the presence of the most common mutations of the KRAS (codons 12 and 13) genes. These regions of interest were amplified using polymerase chain reaction and the reaction product underwent agarose gel electrophoresis against known positive and negative controls to assess the presence and size of the amplified product. Either primary or metastatic tissue was used for the measurements, as a high concordance of the KRAS mutational status between primary and corresponding metastases has been reported (10).
Human sample collection for molecular analysis
To reduce the impact of intra-tumor heterogeneity on molecular analysis, the liver tumor tissue was pulverized using CryoPreTM CP02 (Covaries) and then stored in −80 ℃ refrigerator until being for sequencing. Totally, 232 liver tumor tissues were collected from year July 2018 and to December 2020 in the above participating hospitals. After inclusion and exclusion screenings, a total of 200 eligible subjects were included in our study. According to CPTAC clinical sample collection and procedures, the following criteria were used for the 200 samples: successful extraction of DNA from tumor tissues for whole-exome sequencing. Detailed clinicopathological features are summarized in Table S1. Ethical approval was obtained from the Institutional Review Board of 3 centers. The workflow of multi-omics profiling was showed in Figure 1B.
DNA extraction and whole-exome sequencing (WES)
Genomic DNA was extracted from tumor liver tissues using the DNeasy Blood & Tissue Kit (Qiagen, Venlo, Limburg, The Netherlands) following the manufacturer’s instructions. WES libraries were prepared and captured using the quantified by the Qubit 3.0 (Invitrogen) and NanoDrop 2000 (Thermo Scientific). WES libraries were prepared and captured using the QuarPrep EZ DNA Library Kit and QuarHyb Reagent Kit (Dynasty Gene Biotechnologies) according to manufacturer’s protocol. The DNA library with 150 bp paired-end reads was sequenced with Illumina NovaSeq 6000 System. WES was conducted with an average coverage depth of 597× (range, 500× to 1,000×) for the analysis of somatic and germline mutation landscapes.
Statistical analysis
The categorical variables between the left- and right-sided groups were compared using the c2 test. Overall survival (OS) was defined as time between the day of diagnosis and the day of death or last follow-up. Differences in OS between both groups were assessed via Kaplan-Meier method and the log-rank test, and Cox model adjusting for gender, any treatment prior surgery, tumor sidedness, clinical and pathologic tumor/nodes/metastasis stages, primary tumor differentiation, and metastatic sites presented by maximum size, numbers, and distributions. The effects of selection bias and confounding factors were reduced by calculating the propensity score matching with a ratio of 1:1, and match tolerance less than 0.05. Subgroup analyses were further performed to validate the prognostic value of tumor sidedness in resectable after stratifying by KRAS mutational status.
Additionally, to explore heterogeneity in survival among individuals with different sidedness, we further explored the molecular profiles between the 2 groups. Somatic mutation was performed using the “maftools” R packages. All P values were two-tailed and P values of 0.05 or less were considered statistically significant. Statistical analysis and visualization were performed by using SPSS software (version 22.0; IBM crop, Armonk, NY), GraphPad Prism 9.0.0 and R version 3.6.3 (R Foundation for Statistical Computing, Vienna, Austria, https://www.R-project.org/).
Results
Patient characteristics
Among the 2,434 patients enrolled in the 3 participating centers in Chia, 1,127 (46.3%) patients were excluded due to missing sidedness and molecular data, screen failures, have tumor sited on both the left and right sides, have rectum tumors, or having transverse colon primary tumors (see workflow of study in Figure 1A). This resulted in a total of 1,307 patients included in the present analysis: among them, 755 (57.8%) had left-sided tumors and 552 (42.2%) had right-sided tumors. Patient characteristics by sidedness are included in Table 1. A higher proportion of patients with right-sided tumors were more likely to be female sex (44.0% vs. 32.8%, P<0.001), and had higher tumor stage (90.6% vs. 85.8%, P=0.01) and worse tumor differentiation (30.3% vs. 24.1%, P=0.01) at presentation. Among patients with available molecular marker data, right-sided tumors were more likely to have KRAS (61.6% vs. 35.0%, P<0.001) and BRAF mutations (4.9% vs. 2.5%). In addition, the proportion of patients who were treated with adjuvant systemic treatment (post colectomy) was higher in the right-sided group than in the left-sided group (63.3% vs. 68.9%, P=0.03).
Table 1
| Characteristic | Before PSM | After PSM | |||||
|---|---|---|---|---|---|---|---|
| Left sidedness (N=755) | Right sidedness (N=552) | P value | Left sidedness (N=481) | Right sidedness (N=481) | P value | ||
| Demographic factors | |||||||
| Maternal age, years, mean (SD) | 57.4 (9.5) | 58.8 (9.4) | 0.42 | 57.2 (9.4) | 58.5 (9.2) | 0.37 | |
| Gender, male, n (%) | 507 (67.2) | 309 (56.0) | <0.001 | 297 (61.7) | 282 (58.8) | 0.25 | |
| Body mass index, kg/m2, mean (SD) | 23.6 (6.2) | 23.0 (6.5) | <0.001 | 23.3 (6.6) | 22.8 (6.4) | 0.92 | |
| Neoadjuvant chemotherapy, n (%) | 205 (27.2) | 141 (25.5) | 0.53 | 132 (27.5) | 129 (26.9) | 0.88 | |
| Only chemotherapy | 127 (16.8) | 103 (18.7) | 78 (16.3) | 95 (19.7) | |||
| Combined with bevacizumab | 52 (6.9) | 22 (4.0) | 31 (6.5) | 21 (4.3) | |||
| Combined with cetuximab | 7 (0.9) | 6 (1.1) | 4 (0.9) | 5 (1.0) | |||
| Other treatments | 21 (2.9) | 10 (1.8) | 19 (3.9) | 8 (6.5) | |||
| Adjuvant systemic treatment, n (%) | |||||||
| Post colectomy | 478 (63.3) | 380 (68.9) | 0.03 | 320 (66.5) | 328 (68.2) | 0.58 | |
| Post hepatectomy | 646 (85.6) | 476 (86.3) | 0.73 | 414 (86.1) | 416 (86.4) | 0.85 | |
| Preoperative CEA level, n (%) | <0.99 | <0.99 | |||||
| ≤200 ng/mL | 698 (92.4) | 514 (93.1) | 444 (92.3) | 449 (93.4) | |||
| >200 ng/mL | 57 (7.6) | 38 (6.9) | 37 (7.7) | 32 (6.6) | |||
| Synchronous disease, n (%) | 466 (61.7) | 369 (66.8) | 0.06 | 319 (66.4) | 325 (67.5) | 0.78 | |
| Colorectal site, n (%) | |||||||
| Tumor stage | 0.01 | 0.76 | |||||
| T1–2 | 107 (14.2) | 52 (9.4) | 58 (12.1) | 54 (11.3) | |||
| T3–4 | 648 (85.8) | 500 (90.6) | 423 (87.9) | 427 (88.7) | |||
| Lymph node metastasis | >0.99 | 0.33 | |||||
| Negative | 256 (33.9) | 187 (33.9) | 149 (31.0) | 165 (34.3) | |||
| Positive | 499 (66.1) | 365 (66.1) | 332 (69.0) | 316 (65.7) | |||
| Extrahepatic metastasis | 0.23 | 0.73 | |||||
| No | 636 (84.2) | 451 (81.7) | 399 (82.9) | 394 (82.0) | |||
| Yes | 119 (15.8) | 101 (18.3) | 82 (17.1) | 87 (18.0) | |||
| Tumor differentiation | 0.01 | 0.15 | |||||
| Well/moderate | 573 (75.9) | 385 (69.7) | 344 (71.6) | 365 (75.9) | |||
| Poor | 182 (24.1) | 167 (30.3) | 137 (28.4) | 116 (24.1) | |||
| Metastatic site, n (%) | |||||||
| Largest size, cm | 0.28 | 0.08 | |||||
| ≤3 | 445 (58.9) | 342 (62.0) | 339 (70.5) | 312 (64.9) | |||
| >3 | 310 (41.1) | 210 (38.0) | 142 (29.5) | 169 (37.1) | |||
| Total numbers | 0.83 | 0.93 | |||||
| ≤5 | 621 (82.3) | 457 (82.8) | 395 (82.2) | 397 (82.6) | |||
| >5 | 134 (17.7) | 95 (17.2) | 86 (17.8) | 84 (17.4) | |||
| Distributions | 0.82 | 0.41 | |||||
| Unilobar | 471 (62.4) | 348 (63.0) | 294 (61.2) | 308 (64.0) | |||
| Bilobar | 284 (37.6) | 204 (37.0) | 187 (38.8) | 173 (36.0) | |||
| Molecular features, n (%) | <0.001 | 0.60 | |||||
| KRAS wild-type | 472 (62.5) | 185 (33.5) | 203 (42.2) | 193 (40.1) | |||
| KRAS mutation | 264 (35.0) | 340 (61.6) | 259 (53.8) | 272 (56.6) | |||
| BRAF mutation | 19 (2.5) | 27 (4.9) | 19 (4.0) | 16 (3.3) | |||
CRLM, colorectal liver metastasis; PSM, propensity score matching; SD, standard deviation; CEA, carcinoembryonic antigen; T, tumor.
Prognostic value of primary tumor sidedness
In the overall population (n=1,307), median follow-up was 68 months [95% confidence interval (CI): 16–100 months]. Patients with left-sided tumors had better OS than those with right-sided tumors after accounting for age, sex, prior radiation or adjuvant chemotherapy, tumor/nodes/metastasis stage at presentation, metastatic sites presented by maximum size, numbers and distributions, and molecular features, with a 14.6 lower risk of death [hazard ratio (HR) 1.40, 95% CI: 1.15–1.70, P=0.001] (Figure 2A, Table 2).
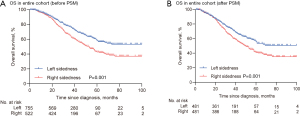
Table 2
| Characteristic | Before PSM | After PSM | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Univariant analysis | Multivariant analysis | Univariant analysis | Multivariant analysis | ||||||||
| HR (95% CI) | P value | HR (95% CI) | P value | HR (95% CI) | P value | HR (95% CI) | P value | ||||
| Neoadjuvant chemotherapy | |||||||||||
| Yes | Reference | Reference | |||||||||
| No | 0.80 (0.64–1.00) | 0.06 | 1.28 (1.01–1.62) | 0.05 | |||||||
| Synchronous metastasis | |||||||||||
| No | Reference | Reference | Reference | Reference | |||||||
| Yes | 1.70 (1.38–2.09) | <0.001 | 1.66 (1.35–2.05) | <0.001 | 1.65 (1.32–2.08) | <0.001 | 1.75 (1.39–2.20) | <0.001 | |||
| Colorectal site | |||||||||||
| Tumor sidedness | |||||||||||
| Left sidedness | Reference | Reference | Reference | Reference | |||||||
| Right sidedness | 1.35 (1.11–1.64) | <0.001 | 1.40 (1.15–1.70) | 0.001 | 1.42 (1.15–1.76) | 0.001 | 1.36 (1.10–1.69) | 0.004 | |||
| Tumor stage | |||||||||||
| T1–2 | Reference | Reference | Reference | ||||||||
| T3–4 | 1.80 (1.26–2.57) | 0.001 | 1.32 (0.90–1.88) | 0.16 | 1.22 (0.96–1.52) | 0.08 | |||||
| Lymph node metastasis | |||||||||||
| Negative | Reference | Reference | Reference | ||||||||
| Positive | 1.69 (1.36–2.11) | <0.001 | 2.02 (1.61–2.54) | <0.001 | 1.50 (1.18–1.89) | 0.001 | 1.33 (1.05–1.69) | 0.02 | |||
| Extrahepatic metastasis | |||||||||||
| No | Reference | Reference | Reference | Reference | |||||||
| Yes | 1.43 (1.13–1.79) | 0.002 | 1.44 (1.16–1.80) | 0.001 | 1.41 (1.10–1.79) | 0.006 | 1.52 (1.19–1.93) | 0.001 | |||
| Tumor differentiation | |||||||||||
| Well/moderate | Reference | Reference | Reference | Reference | |||||||
| Poor | 1.83 (1.50–2.24) | <0.001 | 1.68 (1.36–2.06) | <0.001 | 1.55 (1.25–1.92) | <0.001 | 1.55 (1.25–1.92) | <0.001 | |||
| Metastatic site | |||||||||||
| Largest size | |||||||||||
| ≤3 cm | Reference | Reference | Reference | Reference | |||||||
| >3 cm | 1.57 (1.30–1.91) | <0.001 | 1.36 (1.10–1.67) | 0.004 | 1.66 (1.35–2.05) | <0.001 | 1.36 (1.19–1.69) | 0.004 | |||
| Total numbers | |||||||||||
| ≤5 | Reference | Reference | Reference | ||||||||
| >5 | 2.33 (1.89–2.88) | <0.001 | 2.02 (1.61–2.54) | <0.001 | 2.31 (1.84–2.89 | <0.001 | 2.01 (1.57–2.57) | <0.001 | |||
| Distributions | |||||||||||
| Unilobar | Reference | Reference | Reference | Reference | |||||||
| Bilobar | 1.37 (1.13–1.67) | 0.002 | 0.80 (0.62–1.02) | 0.08 | 1.34 (1.09–1.66) | 0.006 | 0.79 (0.61–1.03) | 0.08 | |||
PSM, propensity score matching; HR, hazard ratio; CI, confidence interval; T, tumor.
Considering that right-sided group was demographically distinct from the left-sided group, and consisted of more advanced cases, we performed propensity score matching of clinical characteristics between the two groups for the analysis of OS in surgery responsiveness. Accordingly, 481 pairs of patients were matched in each group and the baseline characteristics were well-balanced between the two groups (Table 1). The cumulative OS rates at 3-, 5- and 8-year were estimated to be 73.2%, 55.6%, and 50.7% for patients diagnosed with left-sided tumors and 62.7%, 41.0%, and 35.2% for those with right-sided tumors, respectively (P<0.001) (Figure 2B). On the multivariant analysis of propensity score (PS)-matched cohort, left sidedness was associated with improved OS for patients in resected CRLM (HR 1.36, 95% CI: 1.10–1.69, P=0.004) (Table 2).
Subgroup analyses with propensity score matching
Propensity score matching yielded 185, and 256 pairs of patients in both groups in KRAS wild-type and KRAS mutated-type cohorts, respectively (Tables S1,S2). A statistically significant interaction between sidedness and KRAS mutational status were found. Primary tumor sidedness was prognostic among patients with KRAS wild-type tumors, with a 5-year OS for left-sided tumors was 63.1% compared with 39.2% for right-sided tumors (HR 1.71; 95% CI: 1.20–2.45, P=0.003) (Figure 3A, Table 3), but not among patients with KRAS mutated-type tumors (HR 0.88; 95% CI: 0.86–1.16; P=0.36) (Figure 3B, Table 3).
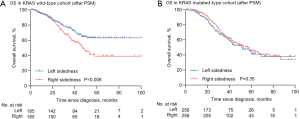
Table 3
| Characteristic | KRAS wild-type tumors | KRAS mutated-type tumors | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Univariate analysis | Multivariate analysis | Univariate analysis | Multivariate analysis | ||||||||
| HR (95% CI) | P value | HR (95% CI) | P value | HR (95% CI) | P value | HR (95% CI) | P value | ||||
| Neoadjuvant chemotherapy | |||||||||||
| Yes | Reference | Reference | Reference | ||||||||
| No | 1.57 (1.09–2.27) | 0.02 | 1.49 (1.00–2.21) | 0.05 | 0.81 (0.58–1.12) | 0.21 | |||||
| Synchronous metastasis | |||||||||||
| No | Reference | Reference | Reference | Reference | |||||||
| Yes | 1.78 (1.20–2.65) | 0.004 | 1.78 (1.16–2.74) | 0.009 | 1.63 (1.22–2.17) | 0.001 | 1.59 (1.19–2.13) | 0.002 | |||
| Colorectal site | |||||||||||
| Tumor sidedness | |||||||||||
| Left sidedness | Reference | Reference | Reference | ||||||||
| Right sidedness | 1.63 (1.15–2.33) | 0.007 | 1.71 (1.20–2.45) | 0.003 | 0.88 (0.66–1.16) | 0.36 | |||||
| Tumor stage | |||||||||||
| T1–2 | Reference | Reference | |||||||||
| T3–4 | 1.50 (0.81–2.78) | 0.20 | 1.52 (0.94–2.47) | 0.09 | |||||||
| Lymph node metastasis | |||||||||||
| Negative | Reference | Reference | Reference | Reference | |||||||
| Positive | 1.59 (1.07–2.36) | 0.02 | 1.56 (1.04–2.33) | 0.03 | 1.76 (1.28–2.41) | <0.001 | 1.54 (1.11–2.13) | 0.009 | |||
| Extrahepatic metastasis | |||||||||||
| No | Reference | Reference | Reference | ||||||||
| Yes | 1.57 (1.04–2.37) | 0.03 | 1.41 (0.92–2.14) | 0.11 | 1.18 (0.85–1.65) | 0.32 | |||||
| Tumor differentiation | |||||||||||
| Well/moderate | Reference | Reference | Reference | Reference | |||||||
| Poor | 1.75 (1.23–2.47) | 0.002 | 1.74 (1.23–2.48) | 0.002 | 1.59 (1.16–2.19) | 0.004 | 1.55 (1.12–2.15) | 0.009 | |||
| Metastatic site | |||||||||||
| Largest size | |||||||||||
| ≤3 cm | Reference | Reference | Reference | Reference | |||||||
| >3 cm | 1.60 (1.13–2.27) | 0.008 | 1.34 (0.90–1.99) | 0.15 | 1.45 (1.09–1.94) | 0.01 | 1.35 (1.00–1.82) | 0.05 | |||
| Total numbers | |||||||||||
| ≤5 | Reference | Reference | Reference | Reference | |||||||
| >5 | 2.22 (1.55–3.18) | <0.001 | 2.86 (1.98–4.15) | <0.001 | 1.53 (1.04–2.25) | 0.03 | 1.33 (0.89–2.00) | 0.17 | |||
| Distributions | |||||||||||
| Unilobar | Reference | Reference | |||||||||
| Bilobar | 1.36 (0.96–1.93) | 0.08 | 1.30 (0.95–1.77) | 0.10 | |||||||
HR, hazard ratio; CI, confidence interval; T, tumor.
The landscape of somatic alternations in left- and right-sided tumors
Efforts to link these clinical differences to specific molecular features have forced on somatic mutations within the coding regions of the genome (11). We performed whole-exome sequencing (WES) on liver tumor tissue of 140 left-sided and 60 right-sided samples to investigate the molecular patterns (Figure 1B; Table S3), obtained from patients who underwent liver resection at Fudan University Shanghai Cancer Center (FUSCC). Mutation frequencies of FBXW7 (9.8% vs. 2.9%), NRAS (6.0% vs. 2.9%), SOX9 (4.5% vs. 0.0%) and CTNNB1 (2.3% vs. 5.9%) were almost identical in both sided tumors, whereas TP53 (58.6% vs. 73.5%, P<0.001), APC (72.9% vs. 94.1%, P<0.001), KRAS (26.3% vs. 32.4%), SMAD4 (18.8% vs. 11.8%), PIK3CA (7.5% vs. 14.7%), and BRAF (3.8% vs. 11.2%) alterations differed between the two groups (Figure 4A), all previously reported as recurrently mutated in CRLM from The Cancer Genome Atlas (TCGA) cohort (12). Copy number variations (CNVs) were successfully determined in 200 samples in our dataset. Frequency affected genes by CNVs in Chinese CRLM patients were HMGB1, BRIC7, FGFR1, PCED1A, SOX17, and SMAD4. The HMGB1 8p11.22 (48.8%) gain was more frequently in the left-sided tumors, and BRIC7 20q13.3 (40.0%) and PCED1A 20p13 (30.0%) loss were more frequently observed in the right-sided tumors (Figure 4B). After adjustment for age, left-sided tumors were also more often harbored SMAD4 mutation but less often carried TP53, APC, and KRAS alterations (Figure 4C). These findings support our hypothesis that tumor sidedness displays unique molecular profiles in CRLM.
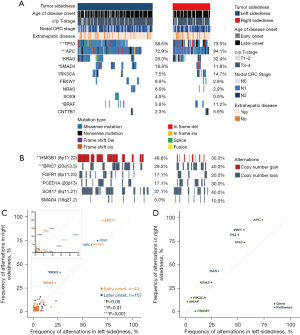
We further investigated the frequencies of oncogenic pathways according to the top 50 mutated genes in each group (Table S4) and found that right-sided tumors more frequently exhibited Wnt, P53, and RAS signaling pathways (Figure 4D).
On univariate analysis, tumor sidedness, the presence of KRAS alterations in the tumor, and BRAF alternations, PIK3CA and SMAD4 alternations were statistically significantly associated with prognosis (Figure S1A). Variables statistically significant on univariate analysis were incorporated into a multivariant model. After adjustment for the statistically significant variables, tumor sidedness was still statistically significantly related with survival (HR 1.32; 95% CI: 1.05–1.64, P=0.02) (Figure S1B). KRAS alterations in the tumor, and BRAF alterations were confirmed as strong prognostic biomarkers associated with poor clinical outcome (Figure S1B). These findings suggest a critical role of KRAS or BRAF alterations that associate with tumor sidedness in treatment responsiveness.
The landscape of germline alternations in left- and right-sided tumors
Germline genomic analysis was performed using an 88-gene panel (left-sided tumors, n=140; right-sided tumors, n=60). This analysis included patients regardless of microsatellite status or known risk factors for CRLM.
Germline pathogenic (P) and likely pathogenic (LP) variant prevalence in all tumor-sided patients was 22.4%. Lynch syndrome (LS) was the most common cancer predisposition syndrome, accounting for of 6.2%. By the clustering of tumor sidedness, the highest mutation prevalence was identified in right-sided cohort with 29.3% harboring an P or LP variant, compared with 15.5% of left-sided patients (P=0.03), and was driven by an enrichment of high-penetrance gene variants (left sidedness =2.1% vs. right sidedness =10.2%, P=0.02). Notably, 68.0% (6 of 9) of these high-penetrance germline mutation carriers harbored P or LP variants in known CRC-associated cancer predisposition genes in the right-sided tumors compared with 24.7% of left-sided tumors, including DNA mismatch repair genes (n=5), and APC (n=1). The distribution of germline variants by gene and penetrance was shown in Figure 5.
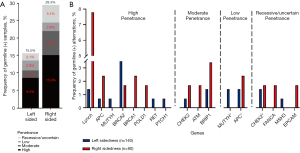
Using matched tumor samples, we interrogated somatic genomic data to assess biallelic inactivation-somatic mutation- at the implicated germline region. Overall, all LP and P germline mutations exhibited biallelic inactivation. Right-sided cohort had the highest rate of biallelic inactivation [43% (right-sidedness) vs. 26% (left-sidedness), P=0.01], suggesting that these germline events were driving CRC carcinogenesis (Figure S2).
Discussion
This study represents a multicenter effort to perform a comprehensive clinical and molecular characterization of left colon cancer versus right colon cancer with liver metastases. We showed that the statistically significant positive prognostic impact of left sidedness was restricted to the patients with KRAS wild-type, while patients with KRAS mutated-type experienced poor outcomes irrespective of the primary tumor sidedness. We further revealed that patients with right-sided tumors displayed unique molecular patterns distinct from left-sided tumors, showing that they more often harbored TP53, APC, KRAS, and BRAF somatic mutations and exhibited higher pathogenic germline variants. Thus, tumor sidedness displays considerable heterogeneity that may associates with their clinical differences.
To our knowledge, this is one of the largest cohorts in the literature for which underlying heterogeneity of primary tumor sidedness has been classified through manual review. Right-sided tumors had a lower incidence, were more prevalent in females, more frequently carried KRAS or BRAF mutations, had a higher tumor stage at presentation, and were associated with worse prognosis than left-sided tumors, which was in line with data previous reported in the literature (13,14). Our findings confirm that tumor sidedness displays considerable heterogeneity that correlates with clinical differences. A possible explanation for these differences is the different embryological origin of the proximal and distal parts of the colon and colorectum. Both parts are joined together at the proximal 2/3 and distal 1/3 of the transverse colon and have different blood supplies, innervations, and lymphatic drainages (15).
Colorectal cancer usually exhibits highly heterogeneity, with molecularly defined subgroups that differ in prognosis (16-19). KRAS (exons 2/3/4) mutations occur in ~30% to 40% of tumors, with increasing interest being used as biologic and molecular markers in the prognostic assessment of patients with metastatic CRC (mCRC) undergoing liver resection (20-22). In our study, we discovered that the clinical outcomes of primary tumor sidedness differed according to the KRAS mutational status. More precisely, primary tumor sidedness was longer a statically significant factor in the multivariate analysis after molecular marker was incorporated into the analysis. This finding is important given that KRAS mutated-type have been shown to be clinically relevant biomarkers associated with the therapeutic benefits of primary tumor sidedness and to have implications for patient outcome. It is clear that proximal and distal CRC should be considered as different clinical entities and tumor sidedness should be considered when making treatment decisions. It should also be included as a stratification factor in future randomized clinical trial, including those assessing impact of treatment sequence, which may also influence long-term outcome. Tumor sidedness is a simple variable, which cannot replace molecular characterization of the tumor but may in part stand as surrogate for complex and still partially understood tumor biology and thus aid clinical decision-making. Note that, in the current analyses, extended RAS mutations beyond KRAS mutation were not included due to limited data availabilities. Given CRLM harboring RAS mutation on exon 3 or 4 behave similarly as KRAS mutation (23) on exon 2 in terms of biological and clinical consequences, the prognostic value of primary tumor location in RAS wild-type CRLM could be potentially more profound.
BRAF (V600E) alterations, harbor by ~10% of tumors, confer poor prognosis (24,25). Concerning BRAF V600E mutants, we did not find any association between primary tumor sidedness and survival in patients with the BRAF V600E mutated CRLM without OS benefit of surgery in KRAS wild-type T BRAF V600E mutated CRLM while the benefits were seen in KRAS wild-type patients. Our results demonstrate that tumor sidedness is a simple variable, which cannot replace molecular characterization of the tumor but may in part stand as surrogate for complex and still partially understood tumor biology and thus aid clinical decision-making.
Given these clinical differences between the two groups, we strived to explore the molecular profiles to describe the considerable heterogeneity. Through a genome-wide characterization of somatic alternations in a series of left and right tumors, we found that right-sided tumors harbored oncogenic mutations, including an enrichment of alternations affecting Wnt, P53 and RAS signaling pathways. These pathways, which appear upregulated in right-sided disease, play a critical role in cellular adhesion and motility, apoptosis, and inflammation, which may in part influence therapy benefits including surgery and chemoradiosensitivity (26). In addition, we showed an overall high frequency of KRAS and BRAF mutations in right-sided tumors, which may reflect the enrichment risk of those patients with worse therapy responsiveness and prognosis. Given the right-sided tumors prevalence of KRAS loss or mutation, we anticipate the treatment benefit to be of widest application among those patients. These findings demonstrate that tumor sidedness displays unique molecular characterization that have important clinical, prognostic, and therapeutic implications.
Our germline analysis revealed that, among patients with right-sided tumors, the prevalence of germline mutations was especially high at 25.6% because of a near doubling of mutation in high-penetrance cancer susceptibility genes. Our integrated germline and somatic analysis helped elucidate the role of these germline variants in CRC carcinogenesis. Biallelic inactivation, resulting from somatic mutation in the tumor, was present in nearly all patients with mutations in a high-penetrance CRLM susceptibility gene and in a higher proportion of right-sided patients, compared with left-sided patients.
There are several limitations to our study. Firstly, we used a retrospectively approach. Thus, our data was inherently flawed by selection and indication bias. Secondly, despite the use of multivariant analysis to enhance intergroup comparison, unidentified biases may have acted in favor of right-sided patients. Thirdly, the proteogenomic characterization of tumor sidedness is necessary to describe the tumor heterogeneity.
Conclusions
This study demonstrates that the prognostic impact of tumor sidedness in surgery responsiveness differs according to the KRAS mutational status. Left-sided CRLM exhibits unique molecular profile distinct from right-sided CRLM that associates with their clinical heterogeneity, which emphasizes the importance of tumor sidedness-based stratification of CRLM, and have critical implications for designing therapeutic strategies.
Acknowledgments
Funding: This work was supported by
Footnote
Reporting Checklist: The authors have completed the STROBE reporting checklist. Available at https://hbsn.amegroups.com/article/view/10.21037/hbsn-22-285/rc
Data Sharing Statement: Available at https://hbsn.amegroups.com/article/view/10.21037/hbsn-22-285/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://hbsn.amegroups.com/article/view/10.21037/hbsn-22-285/coif). The authors reported that this work was supported by National Natural Science Foundation of China (81874182, M-0334), Natural Science Foundation of Shanghai (22ZR1413300), National Science and Technology Major Project (2017ZX10203204-007-004), Shanghai Municipal Health Bureau (201940043), and Shanghai Hospital Development Center (SHDC12019X19). The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by Independent Ethics Committee (IEC) of Fudan University Shanghai Cancer Center, Fifth People’s Hospital of Shanghai Fudan University, and Shanghai Fengxian Central Hospital. Informed consent was obtained from all patients for their data to be used for research.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Zhou H, Liu Z, Wang Y, et al. Colorectal liver metastasis: molecular mechanism and interventional therapy. Signal Transduct Target Ther 2022;7:70. [Crossref] [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin 2020;70:7-30. [Crossref] [PubMed]
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Seligmann JF, Elliott F, Richman S, et al. Clinical and molecular characteristics and treatment outcomes of advanced right-colon, left-colon and rectal cancers: data from 1180 patients in a phase III trial of panitumumab with an extended biomarker panel. Ann Oncol 2020;31:1021-9. [Crossref] [PubMed]
- Alig AHS, Heinemann V, Geissler M, et al. Exact Primary Tumor Location in mCRC: Prognostic Value and Predictive Impact on Anti-EGFR mAb Efficacy. Cancers (Basel) 2022;14:526. [Crossref] [PubMed]
- Boeckx N, Koukakis R, Op de Beeck K, et al. Primary tumor sidedness has an impact on prognosis and treatment outcome in metastatic colorectal cancer: results from two randomized first-line panitumumab studies. Ann Oncol 2017;28:1862-8. [Crossref] [PubMed]
- Petrelli F, Tomasello G, Borgonovo K, et al. Prognostic Survival Associated With Left-Sided vs Right-Sided Colon Cancer: A Systematic Review and Meta-analysis. JAMA Oncol 2017;3:211-9. [Crossref] [PubMed]
- Yin J, Cohen R, Jin Z, et al. Prognostic and Predictive Impact of Primary Tumor Sidedness for Previously Untreated Advanced Colorectal Cancer. J Natl Cancer Inst 2021;113:1705-13. [Crossref] [PubMed]
- Fong Y, Fortner J, Sun RL, et al. Clinical score for predicting recurrence after hepatic resection for metastatic colorectal cancer: analysis of 1001 consecutive cases. Ann Surg 1999;230:309-18; discussion 318-21. [Crossref] [PubMed]
- Vakiani E, Janakiraman M, Shen R, et al. Comparative genomic analysis of primary versus metastatic colorectal carcinomas. J Clin Oncol 2012;30:2956-62. [Crossref] [PubMed]
- Zaidi SH, Harrison TA, Phipps AI, et al. Landscape of somatic single nucleotide variants and indels in colorectal cancer and impact on survival. Nat Commun 2020;11:3644. [Crossref] [PubMed]
- Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012;487:330-7. [Crossref] [PubMed]
- Missiaglia E, Jacobs B, D'Ario G, et al. Distal and proximal colon cancers differ in terms of molecular, pathological, and clinical features. Ann Oncol 2014;25:1995-2001. [Crossref] [PubMed]
- Hsu YL, Lin CC, Jiang JK, et al. Clinicopathological and molecular differences in colorectal cancer according to location. Int J Biol Markers 2019;34:47-53. [Crossref] [PubMed]
- Lee GH, Malietzis G, Askari A, et al. Is right-sided colon cancer different to left-sided colorectal cancer? - a systematic review. Eur J Surg Oncol 2015;41:300-8. [Crossref] [PubMed]
- Guinney J, Dienstmann R, Wang X, et al. The consensus molecular subtypes of colorectal cancer. Nat Med 2015;21:1350-6. [Crossref] [PubMed]
- Vasaikar S, Huang C, Wang X, et al. Proteogenomic Analysis of Human Colon Cancer Reveals New Therapeutic Opportunities. Cell 2019;177:1035-1049.e19. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature 2014;507:315-22. [Crossref] [PubMed]
- Zhang B, Wang J, Wang X, et al. Proteogenomic characterization of human colon and rectal cancer. Nature 2014;513:382-7. [Crossref] [PubMed]
- Mustachio LM, Chelariu-Raicu A, Szekvolgyi L, et al. Targeting KRAS in Cancer: Promising Therapeutic Strategies. Cancers (Basel) 2021;13:1204. [Crossref] [PubMed]
- Porru M, Pompili L, Caruso C, et al. Targeting KRAS in metastatic colorectal cancer: current strategies and emerging opportunities. J Exp Clin Cancer Res 2018;37:57. [Crossref] [PubMed]
- Pfeiffer P, Qvortrup C. KRAS(G12C) inhibition in colorectal cancer. Lancet Oncol 2022;23:10-1. [Crossref] [PubMed]
- Allegra CJ, Rumble RB, Hamilton SR, et al. Extended RAS Gene Mutation Testing in Metastatic Colorectal Carcinoma to Predict Response to Anti-Epidermal Growth Factor Receptor Monoclonal Antibody Therapy: American Society of Clinical Oncology Provisional Clinical Opinion Update 2015. J Clin Oncol 2016;34:179-85. [Crossref] [PubMed]
- Vanni I, Tanda ET, Spagnolo F, et al. The Current State of Molecular Testing in the BRAF-Mutated Melanoma Landscape. Front Mol Biosci 2020;7:113. [Crossref] [PubMed]
- Margonis GA, Buettner S, Andreatos N, et al. Association of BRAF Mutations With Survival and Recurrence in Surgically Treated Patients With Metastatic Colorectal Liver Cancer. JAMA Surg 2018;153:e180996. [Crossref] [PubMed]
- Kirzin S, Marisa L, Guimbaud R, et al. Sporadic early-onset colorectal cancer is a specific sub-type of cancer: a morphological, molecular and genetics study. PLoS One 2014;9:e103159. [Crossref] [PubMed]


