
A literature review of genetics and epigenetics of HCV-related hepatocellular carcinoma: translational impact
Introduction
The occurrence of hepatocellular carcinoma (HCC) has notably increased in recent decades, positioning it as the fifth most prevalent form of cancer worldwide (1,2). HCC is a heterogeneous disease, with the underlying causes mostly due to persistent liver damage and cirrhosis (3). The prognosis and survival rates of HCC are unfavourable, and therapeutic options are still limited. HCC now ranks as the second highest cause of cancer-related mortality among males, resulting in about one million fatalities on a yearly basis (2).
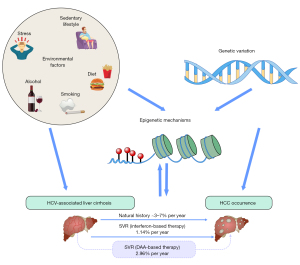
The hepatitis C virus (HCV) continues to pose a significant challenge to public health, affecting a global population of over 71 million individuals and resulting in an annual incidence of 3–4 million new infections (4). HCV is still a leading cause of HCC globally, with a reported average incidence of 1–4% per year in HCV-related cirrhosis (5). A recent meta-analysis has shown that there is no statistically significant difference in the estimated incidence of HCC occurrence after sustained virological response (SVR) between treatments based on interferon (IFN) and those based on direct-acting antivirals (DAAs) {1.14/100 patient-years (py) [95% confidence interval (CI): 0.86–1.52] and 2.96/100 py (95% CI: 1.76–4.96) in IFN and DAA studies, respectively} (6). While treatment for HCV markedly improved the outcome of HCV eradication, increased risk for HCC development is maintained even after SVR following DAA therapy (7). It is anticipated that there will be a gradual rise in the number of patients who have achieved SVR after DAA treatment in the next decade (8). However, the burden of HCC linked with HCV is projected to persist in the years to come; thus, understanding the mechanisms involved and patient risk factors will help physicians target these patients and alleviate the HCC burden.
The mechanism by which HCV causes liver cancer is not completely understood. The pathogenic process is intricate and multifaceted, encompassing a dynamic interaction between host factors, such as genetic predisposition, and environmental and viral factors. Some other major risk factors involved in the HCC development include ethinicity, diabetes, exposure to aflatoxin, smoking, and heavy alcohol use and co-existing of metabolic-associated fatty liver disease (MAFLD) or chronic hepatitis B infection. Emerging evidence suggests that numerous genetic and epigenetic mechanisms play a key in HCC pathogenesis (9). Furthermore, HCC occurs even long after the eradication of HCV, indicating the implications of the persistent genetic and epigenetic alterations in HCV-related hepatocarcinogenesis (Figure 1). Here, we provide a comprehensive and updated review of genetics and epigenetic processes involved in HCV-related HCC. We also discuss the potential translational implications, including novel biomarkers and drugs for treatment. We present this article in accordance with the Narrative Review reporting checklist (available at https://hbsn.amegroups.com/article/view/10.21037/hbsn-23-562/rc).
Methods
The search strategy is described in Table 1.
Table 1
| Items | Specification |
|---|---|
| Date of search | June 2023 |
| Databases searched | PubMed and Embase |
| Search terms used | “Hepatocellular carcinoma”, “Hepatitis C”, “Genetic”, “Epigenetic”, “SNP”, “methylation”, “Histone” AND “GWAS” |
| Timeframe | The search encompassed articles published from 2000 to June 2023 |
| Inclusion criteria | Articles in English language. The search strategy aimed to identify relevant studies for an in-depth review, providing valuable insights into the relationship between genetic, epigenetic and HCV related HCC, and advancements in diagnosis and therapeutic approaches |
| Selection process | Z.P. conducted the selection, and all authors revised the manuscript to identify potential studies based on their expertise |
SNP, single nucleotide polymorphism; GWAS, genome-wide association studies; HCV, hepatitis C virus; HCC, hepatocellular carcinoma.
Genetics: HCV-related HCC
Two genome-wide association studies (GWAS) of HCV-HCC conducted in Japan identified two risk variants in the 5' flanking region of the major histocompatibility complex class I-related chain A (MICA) on 6p21.33 (rs2596542) and DEP domain containing 5, GATOR1 subcomplex subunit (DEPDC5) rs1012068 (10,11). Further investigation revealed that the MICA variation does not exhibit an association with HCC susceptibility. However, it does demonstrate a correlation between heightened severity of fibrosis and accelerated fibrosis progression. This suggests that the MICA variant is involved in the susceptibility to advanced fibrosis, which indirectly impacts the likelihood of developing HCC. Mechanistically, the hepatic and serum MICA expression was attenuated in advanced fibrosis stages and in patients harbouring the MICA risk allele via transforming growth factor-β1 (TGF-β1)-dependent mechanisms (12). In this study, no significant association was seen between the DEPDC5 variation and the incidence of either fibrosis or HCC (12). A subsequent GWAS study and large candidate gene studies discovered a specific genetic variation within the tolloid-like protease 1 gene that is associated with the advancement of fibrosis and HCC progress following SVR (13-15), but it has no role in patients with MAFLD (16).
In the context of candidate gene research, a recent systematic review has shown that 137 distinct genes have been associated with HCV-HCC. Among these genetic factors, the genes IFN-λ3/IFN-λ4, tumour necrosis factor-alpha (TNF-α), and Patatin-like phospholipase domain-containing protein 3 (PNPLA3) exhibited the most robust and consistent associations (17).
Genetic variation in the IFNL3/IFNL4 region was originally identified in GWAS studies as a genetic determinant of HCV clearance. Subsequent investigations have expanded upon these initial discoveries and provided more evidence to support the strong association between the IFNL3/IFNL4 region and the occurrence of hepatic inflammation and fibrosis in individuals diagnosed with chronic hepatitis C (CHC), as well as other liver diseases of various causes (18-22) and lung fibrosis (23). However, the role of these variants in HCC risk and recurrence after therapy is less clear, with some mixed results reported. Early study failed to discern any association between the IFNL3/IFNL4 genotype and the development of HCC in patients with HCV cirrhosis (24). Conversely, other studies found that HCC was more prevalent and likely to occur at a younger age in patients harbouring the non-responder genotype compared to those with the CC genotype (25,26). Furthermore, another study involving patients with HCV indicated that those with the TT genotype had a greater susceptibility to HCC development and displayed elevated levels of alpha-fetoprotein in comparison to those with the responder genotype (27). In regard to HCC recurrence, a Japanese study suggested that patients with CC genotype were significantly more likely to have HCC recurrence after curative therapy (28). Collectively, these findings suggest that patients harbouring the IFNL3/IFNL4 genotype non-responder genotype have diminished immune responses, with an augmented risk of tumour escape. In this context, recent studies showed decreased hepatic and circulating immune cells in patients with the IFNL3/IFNL4 genotype non-responder genotype (20,29). Similarly, multiple studies demonstrated an association between TNF-α single nucleotide polymorphisms (SNPs) and HCV-related HCC (30,31).
The rs7574865 variant in the signal transducers and activators of transcription (STAT4) gene, which is another variant related to immunity, has been suggested to be linked with various phenotypes of hepatitis B virus (HBV) infection. This includes spontaneous HBV clearance, response to IFN-α-based therapy, and the risk of developing fibrosis and HCC (32,33). In contrast, this variant was not associated with any of these phenotypes in patients with HCV, suggesting a differential role of immune responses and related genetic variants between these two viral hepatitis (34,35).
Genetic risk variants for fatty liver disease have been reported to influence the risk of fibrosis progression and HCC across various aetiologies, including HCV. PNPLA3 p.I148M, transmembrane 6 and superfamily member 2 (TM6SF2) p.E167K were reported to be associated with diverse roles across various liver diseases (36). Recent research showed that there is no significant association between the PNPLA3 rs738409 variant, the TM6SF2 rs58542926 variant, or the combined presence of at-risk alleles and the incidence of HCC in individuals with cirrhosis caused by HCV infection (35). In 110 patients with HCV-HCC, the coexistence of the PNPLA3 and HSD17B13 risk alleles was an independent risk factor for HCC (37). The role of other genetic variants associated with hepatic fat accumulation and metabolic steatohepatitis in HCV-HCC is yet to be characterised (38-40).
Some of these genetic variants have been implicated in the risk of HCC in those with SVR induced by pegylated interferon alpha-2a/ribavirin (PegIFN/RBV) or DAA. A study conducted on a cohort of 1,118 Taiwanese patients with a median duration of 60 months diagnosed with HCV infection aimed to investigate the potential relationship between IFNL3/IFNL4 genotype and the development of HCC. The findings revealed no significant relationship between IFNL3/IFNL4 genotype and HCC in individuals who achieved SVR through treatment with PegIFN/RBV. However, individuals with the non-responder genotype (CT/TT) had a higher risk of developing HCC compared to those with the responder genotype (CC) among individuals who did not achieve SVR (41). In another investigation including a cohort of 493 individuals from Egypt, 70 were classified as healthy controls, while 252 were patients with HCV who received DAA treatment. Among the patients with HCV, 65 individuals developed HCC. The study results revealed no statistically significant correlation between the incidence of HCC and the carriage of the IFNL3/IFNL4 rs12979860 SNP or the TLR4 rs4986791 variation (42).
Furthermore, a comprehensive analysis conducted on a cohort of 3,771 Japanese individuals diagnosed with HCV who successfully achieved SVR revealed a noteworthy correlation between PNPLA3 and HLA-DQB1 polymorphisms with HCC development subsequent to SVR. This association was especially pronounced among individuals who were not classified as obese, which included a median duration of observation spanning 41 months (43). A recent investigation involving 290 Japanese patients with CHC who had not previously developed HCC and had successfully eliminated the HCV through DAA treatment revealed that PNPLA3 GG genotype individuals were more at risk in HCC experience, regardless of their age, gender, FIB-4 index, serum albumin levels, alanine aminotransferase levels, and alpha-fetoprotein levels at 12 weeks post-DAA treatment. The adjusted hazard ratio was 2.57, with a 95% CI of 1.01–6.91 and a P value of 0.048 (44).
Similarly, a variant in MBOAT7, a protein in the Lands cycle, has been associated with liver injury in MAFLD, HBV, and HCV, but not HCC (45-47). Another study of 200 patients who underwent DAA treatment for CHC with advanced fibrosis (F3–F4) showed that IFNL3-TT genotype was associated with HCC development after DAA therapy [odds ratio (OR) 4.728, 95% CI: 1.222–18.297, P=0.024], but not other variants PNPLA3 (rs738409), TM6SF2 (rs58542926) or MBOAT7 (rs641738) (48). Lastly, a study of 509 consecutive Italian patients with HCV cirrhosis, with a median follow-up of 43 [3–57] months after DAA start, showed a genetic risk score combining variants in PNPLA3, MBOAT7, TM6SF2, and GCKR, but each of these variations was exclusively linked to the development of de novo HCC, regardless of traditional risk variables such as the severity of liver disease (49). Whether, the genetic contribution to HCC would differ after SVR via Peg-IFN/RBV or DAA is yet to be determined.
Epigenetics
Epigenetics studies the heritable and acquired modifications in gene function and expression that determine phenotype, independent of alteration in the DNA sequence, that are frequently identified in various cancer types, including HCC (50). Classically, epigenetic modifications are grouped into three main classes: DNA methylation, histone modifications (acetylation and methylation), and RNA-based mechanisms [microRNAs (miRNAs) and non-coding RNAs (ncRNAs)] (51).
Epigenetic modulators refer to a collection of genes that are situated upstream of the epigenetic modifiers and mediators (52). In cancers, external (carcinogens or viral infection) or internal (ageing or metabolic comorbidities) factors drive the neoplastic propensity imposed on epigenetic modulators (52). Some remarkable examples of epigenetic modulators are oncogenic RAS, nuclear factor-κB, and tumour suppressor protein p53 (52).
Emerging evidence suggests that HCV infection induces a substantial remodelling of cancer-related pathways via epigenetic interference (53). The existence of HCV RNA or/and expression of the viral proteins in the cells is pivotal in inducing these epigenetic alterations that subsequently reprogram host gene expression (53). For instance, HCV infection promotes reactive oxygen species generation and S-adenosyl-homocysteine, causing abnormal hypermethylation and chromatin modifications of the antitumour genes that potentiate HCC progression, which are thereby implicated in the initiation of carcinogenesis, progression, and metastasis development (54-56). The HCV-induced epigenetic signature has been found to be long-term, sustaining even after infection cure, which could be implicated in the pathogenesis of the persistence of HCC risk following HCV viral cure (57).
DNA methylation
DNA methylation is a crucial epigenetic alteration that serves as the primary mechanism for regulating gene expression. This refers to the occurrence of methylation in cytosine bases that are concentrated inside regions known as CpG-islands (50,58). The pathophysiology of diseases, including cancers, is significantly influenced by abnormal DNA methylation patterns seen in gene promoters. These patterns, characterized by either hypermethylation or hypomethylation, may lead to the transcriptional suppression or activation of tumour-suppressor or oncogenic genes, respectively (50,58).
Significant alterations in DNA methylation have been observed in HCC in recent studies using genome-wide DNA methylation profiling techniques (59-61). The activation of an innate immune response mediated by natural killer cells was seen in chimeric mice with humanised livers upon HCV infection. Subsequently, this was accompanied by a genome-wide alteration of DNA methylation in human hepatocytes (62). Similarly, in humans, HCV can influence the methylation patterns of CpG islands, particularly in genes associated with DNA mismatch repair (63) and/or the control of the cell cycle (64).
Notably, the HCV-induced DNA methylation changes were distinct from that of HBV. A comprehensive genome-wide analysis of DNA methylation patterns using the Illumina HumanMethylation450 BeadChip revealed distinct differences in methylation profiles between HCC caused by HBV and HCC caused by HCV. Specifically, a majority of the CpG sites that exhibited differential methylation were shown to be hypermethylated in HCV-HCC in comparison to HBV-HCC (65).
Histone modification
Histone modifications are dynamic forms of epigenetic change that serve as points of regulation for the transcriptional machinery and chromatin compaction (66). Histones possess N-terminal tails that extend beyond the nucleosomes and experience a diverse array of post-translational modifications. The acetylation and methylation of histones are among the most intensively researched post-translational changes. Acetylation of histones is generally recognized as an activating change, while methylation of histones may have either activating or repressive effects.
Extensive research has been conducted on investigating the histone alterations signatures associated with HCC development. Currently, there is convincing evidence that HCV induces a substantial remodelling in host chromatin structure (67) that remains even after achieving SVR using DAAs (67).
New research using chromatin immunoprecipitation followed by a next-generation sequencing (ChIP-seq) technique unveiled that HCV infection elicits significant alterations in the epigenetic landscape throughout the whole genome (57). The alterations may be categorized into an 8-gene signature that exhibits an association with the development of HCC. These changes include activation of the oncogenic network and suppression of the tumour-suppressor genes, and these changes are likely implicated in the pathogenesis of HCV-induced hepatocarcinogenesis (68).
Another study showed similar findings that HCV induces specific epigenetic changes, namely genome-wide changes in H3K27ac, which persist after SVR and are associated with changes in the expression of mRNAs and proteins and HCC risk (67). A comprehensive analysis of several epigenetic alterations in a cell line infected with HCV was recently conducted. Specifically, they examined four histone modifications (H3K4me3, H3K4me1, H3K27ac, and H3K27me3), open chromatin using ATAC-seq, and genome-wide DNA methylation. Additionally, they investigated the effects of HCV eradication by treatment with either IFN-α or a DAA agent in these experimental models (69). This work demonstrated that HCV infection substantially influences the epigenome, with shared epigenetic changes with IFN-α effect. The majority of these changes induced by HCV remain as ‘epigenomic signature’ following viral cure (69).
Non-coding RNAs
Two distinct categories of ncRNAs, namely miRNAs and long non-coding RNAs (lncRNAs), have been extensively investigated and are recognized as significant contributors to epigenetic regulation. ncRNAs have been seen to exert regulatory effects on gene expression at both the transcriptional and post-transcriptional levels. Their involvement in the control of several cancers, including HCC, is well-established and considered significant (50).
miRNAs are a class of ncRNAs that consist of single-stranded molecules ranging from 19 to 22 nucleotides in length. Their primary mechanism of action involves the suppression of target mRNA by binding to complementary sequences. Therefore, they play a crucial role in regulating gene expression at the post-transcriptional stage (70).
miRNAs have been identified as regulators of many biological pathways implicated in the development of HCC (71). For example, miR-122 is a prominent miRNA mostly expressed in the liver, constituting around 70% of the total miRNA population in this organ. A study has reported that miR-122, which acts as a tumour-suppressor gene, is implicated in HCV-related HCC, which has been confirmed by a recent meta-analysis (72). A further investigation revealed that levels of blood miRNA-224 were notably higher in patients with HCV-HCC in comparison to both cirrhotic patients and those in good health who served as control subjects (73). Another study observed a notably increased expression level of serum miRNA-27a in patients with HCV-HCC in comparison to those with cirrhosis. Furthermore, these raised levels were shown to be associated with distant metastasis, Child-Pugh grade, and lymph node metastasis (74).
LncRNAs represent a class of RNA molecules that exceed a length of 200 bases. Although this group has no protein-coding function, they act via cis- or trans-regulation and regulate diverse functions, including mRNA stability, chromatin accessibility, and protein activity, and are implicated in multiple aspects of the progression of various cancers (75,76). A study demonstrates alterations in lncRNAs in HCC. Firstly, HCV induced down-regulation of a lncRNA, Linc-Pint, which enhances virus replication and liver disease progression through increasing lipogenesis (77). Secondly, the blood expression levels of lnc-HOX antisense intergenic RNA were elevated in patients with HCV-HCC treated with DAAs (78). Lastly, the expression level of lnc-taurine-upregulated gene 1 and lnc-cancer susceptibility candidate 2 in polymorphonuclear leucocytes were demonstrated to be upregulated and downregulated in HCV-HCC compared to patients with HCV and healthy controls, respectively (79).
Genetics and epigenetic biomarkers and therapeutic targets
Biomarkers
Genetic markers and epigenetic modifications have the potential to function as innovative biomarkers for the risk stratification and early detection of HCC enabling more effective HCC screening prioritization of patients with HCC for therapies. The process of stratifying patients based on their genetic risk level allows for the identification of individuals with an inherited greater risk of developing HCC. This enables the prioritization of early interventions and therapies for those who are most in need, with the goal of preventing the advancement of the disease. Additionally, the genetic and epigenetic biomarkers can be incorporated into novel risk scores with other clinical variables (80-84), particularly metabolic risk factors that are well implicated in the risk of HCC in patients with cured HCV (85). For instance, a recent meta-analysis suggested the potential utility of lncRNA as a diagnostic biomarker for HCC (86). This study indicates that the co-assessment of several lncRNAs, namely HULC, Linc00152, RP11-160H22.5, XLOC_014,172, LOC149086, uc001ncr, and AX800134 yields a receiver operating characteristic (ROC) curve with improved specificity and sensitivity compared to the evaluation of a single lncRNA. Specifically, the area under the curve (AUC) for the combined assessment is 0.94, while the AUC for a single lncRNA is 0.902.
The potential use of particular miRNAs from the sera of patients with HCC as a biomarker has been proposed (87). The study has shown the presence of elevated levels of miR-133a, miR-192, miR-143, miR-29a, miR-145, miR-29c, and miR-505 in individuals diagnosed with HCC (87). In addition, it has been shown that miR-219-5p has a role in facilitating metastasis in HCC and is correlated with a worse prognosis for patients (88). Consequently, miRNAs might be considered as a potential prognostic indicator for HCC and its progression (88).
As previously mentioned, many observations of DNA methylation patterns in the genes exhibit a correlation with dysregulated signalling pathways important for HCC development. Therefore, a comprehensive investigation of the methylome has the potential to uncover hitherto unidentified biomarkers for HCC.
Genetics and epigenetic therapeutic targets
The only available curative therapy to treat patients who develop HCC is surgery (resection or transplantation); nevertheless, the vast majority of patients not being candidates for this option, and thus, other treatment modalities are needed. Tumour ablation using radiofrequency and microwave, as well as transarterial chemoembolization, represents an alternative treatment option for HCC. Additionally, sorafenib is considered the recommended treatment choice for individuals who are deemed ineligible for surgical intervention, and there are now more pharmaceutical options for HCC, such as lenvatinib, a multikinase inhibitor, and nivolumab, a programmed cell death protein-1 immune checkpoint inhibitor. Atezolizumab plus bevacizumab, sorafenib, and lenvatinib is recommended as a first-line option by most guidelines (89). Although these drugs represent a significant step forward in increasing the survival of patients, the response rate is still substantially suboptimal. Given the current shortage in treatment modalities for HCC, innovative therapeutic strategies for HCC are urgently needed.
Genetics can guide novel therapeutic approaches (90). Analyses of various pharmaceutical pipelines found that targets with a genetic link to disease have more than double the chance of success in clinical trials and obtaining regulatory approval compared to those lacking such a link (91,92). Therefore, insights from the GWAS studies for HCC could potentially offer an additional avenue for priortising drug targets and safety, both key requirements in drug discovery. Genetics can also inform drug repurposing, accelerating the drug development process. Some notable examples are TGF-β1 at 19q13.2, which is a targeted drug used to treat rheumatoid arthritis and is also linked to an elevated risk of colorectal cancer (93). Another significant genetic locus is the vitamin D receptor at 12q13.11, which is a drug target for treating osteoporosis and a risk locus for prostate cancer (94). Lastly, the tyrosinase gene at 11q14.3, the target of an approved drug used in the treatment of skin disorders (95), is also associated with an increased risk of developing melanoma (95), squamous cell carcinoma (96), and basal cell carcinoma (97).
In addition, the plasticity and reversibility of the epigenome may provide a unique opportunity to intervene and modulate the unfavourable epigenetic states for preventing and treating HCC. Inhibiting the function of epigenetic modulators, for instance, epigenetic enzymes (such as histone acetyltransferases), can be exploited as a plausible therapeutic approach for attaining pharmacological restoration of the normal epigenetic states (98). In recent years, epidrugs have gained significant attention and have been widely tested and utilised, with many candidates currently under clinical trials for therapy of various malignancies (98). In this context, inhibitors for DNA methyltransferase (DNMT) and histone deacetylase (HDAC) are currently being experimentally tested for treatment of HCC (99). Reminostat, an oral pan-HDAC inhibitor, efficiently enhances the sensitivity to sorafenib in the treatment of patients with advanced HCC (99). In a similar way, the concurrent administration of quisinostat, an additional HDAC inhibitor, and sorafenib showed a synergistic effect in reducing the development of HCC (100). CM-272, a novel first-class small-molecule dual histone methyltransferase G9a and DNMT1 inhibitor, has a suppressive effect on experimental HCC models (101). Recent research has shown comparable epigenetic and transcriptome alterations connected to cancer between CHC and MAFLD (102). The study findings demonstrate that the pharmacological suppression of bromodomain 4, a chromatin reader, effectively mitigated the course of liver disease and hepatocarcinogenesis. This beneficial effect was achieved via epigenetic and transcriptional reprogramming (102).
Conclusions
The incidence of HCC has been strikingly rising worldwide, and the treatment options are still limited. It has therefore become the fastest-increasing cause of cancer-related mortality. Even in the current era of efficient HCV DAA therapy, with a very high response rate, it has become obvious that the risk for HCC occurrence persists after HCV eradication. The identification of genetic and epigenetic mechanisms that are implicated in maintaining HCC risk following HCV cure provides a unique opportunity to envisage diagnostic tools and algorithms to predict the risk of HCC occurrence in this growing population and pave the way for novel therapeutic strategies, targeting the hypothetically reversible oncogenic process of epigenetics leading to cancer after HCV cure, unlike the irreversible mutations. In addition, this approach can offer a new path for HCC prevention, including after HCV-induced liver damage occurs. Future studies are required to reveal specific epigenetic upstream and modulators and identify druggable targets that regulate hub pathways induced by HCV, leading to dysregulation of the epigenome. Leveraging these mechanisms can ultimately lead to the identification and development of HCV-aetiology-specific personalised intervention approaches for HCC management and prevention. The application of artificial intelligence can be a possible solution. The forthcoming wave of research is expected to incorporate the integration of multilayered information through multi-omics data analysis and artificial intelligence methods. This approach will enable the precise stratification of patients, leading to more personalized management approaches.
Acknowledgments
Funding: This work was supported by
Footnote
Reporting Checklist: The authors have completed the Narrative Review reporting checklist. Available at https://hbsn.amegroups.com/article/view/10.21037/hbsn-23-562/rc
Peer Review File: Available at https://hbsn.amegroups.com/article/view/10.21037/hbsn-23-562/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://hbsn.amegroups.com/article/view/10.21037/hbsn-23-562/coif). Y.M. serves as the Editor-in-Chief of HepatoBiliary Surgery and Nutrition. M.E. serves as an unpaid editorial board member of HepatoBiliary Surgery and Nutrition. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- El‐Serag HB. Epidemiology of hepatocellular carcinoma. In: Arias IM, Alter HJ, Boyer JL, et al. editors. The liver: Biology and pathobiology. Hoboken, NJ: Wiley; 2020:758-772.
- Dragani TA. Risk of HCC: genetic heterogeneity and complex genetics. J Hepatol 2010;52:252-7. [Crossref] [PubMed]
- Progress report on access to hepatitis C treatment: focus on overcoming barriers in low-and middle-income countries. World Health Organization; 2018.
- Organization WH. WHO urges countries to invest in eliminating hepatitis. 2019. In:2019.
- Waziry R, Hajarizadeh B, Grebely J, et al. Hepatocellular carcinoma risk following direct-acting antiviral HCV therapy: A systematic review, meta-analyses, and meta-regression. J Hepatol 2017;67:1204-12. [Crossref] [PubMed]
- Shiha G, Waked I, Soliman R, et al. GES: A validated simple score to predict the risk of HCC in patients with HCV-GT4-associated advanced liver fibrosis after oral antivirals. Liver Int 2020;40:2828-33. [Crossref] [PubMed]
- Lynch EN, Russo FP. Outcomes and Follow-Up after Hepatitis C Eradication with Direct-Acting Antivirals. J Clin Med 2023;12:2195. [Crossref] [PubMed]
- Eslam M, George J. Genome-Wide Association Studies and Hepatitis C: Harvesting the Benefits of the Genomic Revolution. Semin Liver Dis 2015;35:402-20. [Crossref] [PubMed]
- Kumar V, Kato N, Urabe Y, et al. Genome-wide association study identifies a susceptibility locus for HCV-induced hepatocellular carcinoma. Nat Genet 2011;43:455-8. [Crossref] [PubMed]
- Miki D, Ochi H, Hayes CN, et al. Variation in the DEPDC5 locus is associated with progression to hepatocellular carcinoma in chronic hepatitis C virus carriers. Nat Genet 2011;43:797-800. [Crossref] [PubMed]
- Sharkawy RE, Bayoumi A, Metwally M, et al. A variant in the MICA gene is associated with liver fibrosis progression in chronic hepatitis C through TGF-β1 dependent mechanisms. Sci Rep 2019;9:1439. [Crossref] [PubMed]
- John M, Metwally M, Mangia A, et al. TLL1 rs17047200 Increases the Risk of Fibrosis Progression in Caucasian Patients With Chronic Hepatitis C. Gastroenterology 2017;153:1448-9. [Crossref] [PubMed]
- Matsuura K, Sawai H, Ikeo K, et al. Genome-Wide Association Study Identifies TLL1 Variant Associated With Development of Hepatocellular Carcinoma After Eradication of Hepatitis C Virus Infection. Gastroenterology 2017;152:1383-94. [Crossref] [PubMed]
- Huang CF, Yeh ML, Huang CI, et al. Tolloid-like 1 genetic variants determine fibrosis regression in chronic hepatitis C patients with curative antivirals. Sci Rep 2018;8:15058. [Crossref] [PubMed]
- Bayoumi A, Jalil I, Metwally M, et al. Genetic variation in the TLL1 gene is not associated with fibrosis in patients with metabolic associated fatty liver disease. PLoS One 2020;15:e0243590. [Crossref] [PubMed]
- Walker AJ, Peacock CJ, Pedergnana V, et al. Host genetic factors associated with hepatocellular carcinoma in patients with hepatitis C virus infection: A systematic review. J Viral Hepat 2018;25:442-56. [Crossref] [PubMed]
- Eslam M, Ahlenstiel G, George J. Interferon Lambda and Liver Fibrosis. J Interferon Cytokine Res 2019;39:627-35. [Crossref] [PubMed]
- Eslam M, Hashem AM, Leung R, et al. Interferon-λ rs12979860 genotype and liver fibrosis in viral and non-viral chronic liver disease. Nat Commun 2015;6:6422. [Crossref] [PubMed]
- Eslam M, McLeod D, Kelaeng KS, et al. IFN-λ3, not IFN-λ4, likely mediates IFNL3-IFNL4 haplotype-dependent hepatic inflammation and fibrosis. Nat Genet 2017;49:795-800. [Crossref] [PubMed]
- Eslam M, Hashem AM, Romero-Gomez M, et al. FibroGENE: A gene-based model for staging liver fibrosis. J Hepatol 2016;64:390-8. [Crossref] [PubMed]
- Eslam M, Leung R, Romero-Gomez M, et al. IFNL3 polymorphisms predict response to therapy in chronic hepatitis C genotype 2/3 infection. J Hepatol 2014;61:235-41. [Crossref] [PubMed]
- Metwally M, Thabet K, Bayoumi A, et al. IFNL3 genotype is associated with pulmonary fibrosis in patients with systemic sclerosis. Sci Rep 2019;9:14834. [Crossref] [PubMed]
- Bochud PY, Bibert S, Kutalik Z, et al. IL28B alleles associated with poor hepatitis C virus (HCV) clearance protect against inflammation and fibrosis in patients infected with non-1 HCV genotypes. Hepatology 2012;55:384-94. [Crossref] [PubMed]
- Sato M, Kato N, Tateishi R, et al. IL28B minor allele is associated with a younger age of onset of hepatocellular carcinoma in patients with chronic hepatitis C virus infection. J Gastroenterol 2014;49:748-54. [Crossref] [PubMed]
- Fabris C, Falleti E, Cussigh A, et al. IL-28B rs12979860 C/T allele distribution in patients with liver cirrhosis: role in the course of chronic viral hepatitis and the development of HCC. J Hepatol 2011;54:716-22. [Crossref] [PubMed]
- Eurich D, Boas-Knoop S, Bahra M, et al. Role of IL28B polymorphism in the development of hepatitis C virus-induced hepatocellular carcinoma, graft fibrosis, and posttransplant antiviral therapy. Transplantation 2012;93:644-9. [Crossref] [PubMed]
- Hodo Y, Honda M, Tanaka A, et al. Association of interleukin-28B genotype and hepatocellular carcinoma recurrence in patients with chronic hepatitis C. Clin Cancer Res 2013;19:1827-37. [Crossref] [PubMed]
- O'Connor KS, Read SA, Wang M, et al. IFNL3/4 genotype is associated with altered immune cell populations in peripheral blood in chronic hepatitis C infection. Genes Immun 2016;17:328-34. [Crossref] [PubMed]
- Tarhuni A, Guyot E, Rufat P, et al. Impact of cytokine gene variants on the prediction and prognosis of hepatocellular carcinoma in patients with cirrhosis. J Hepatol 2014;61:342-50. [Crossref] [PubMed]
- Talaat RM, Esmail AA, Elwakil R, et al. Tumor necrosis factor-alpha -308G/A polymorphism and risk of hepatocellular carcinoma in hepatitis C virus-infected patients. Chin J Cancer 2012;31:29-35. [PubMed]
- Jiang DK, Sun J, Cao G, et al. Genetic variants in STAT4 and HLA-DQ genes confer risk of hepatitis B virus-related hepatocellular carcinoma. Nat Genet 2013;45:72-5. [Crossref] [PubMed]
- El Sharkawy R, Thabet K, Lampertico P, et al. A STAT4 variant increases liver fibrosis risk in Caucasian patients with chronic hepatitis B. Aliment Pharmacol Ther 2018;48:564-73. [Crossref] [PubMed]
- Asimakopoulos A, Mangia A, Dore GJ, et al. Polymorphisms in STAT4 are not associated with treatment response and spontaneous clearance of hepatitis C virus in europeans. Hepatology 2016;64:2264-5. [Crossref] [PubMed]
- Yang J, Trépo E, Nahon P, et al. PNPLA3 and TM6SF2 variants as risk factors of hepatocellular carcinoma across various etiologies and severity of underlying liver diseases. Int J Cancer 2019;144:533-44. [Crossref] [PubMed]
- Eslam M, Mangia A, Berg T, et al. Diverse impacts of the rs58542926 E167K variant in TM6SF2 on viral and metabolic liver disease phenotypes. Hepatology 2016;64:34-46. [Crossref] [PubMed]
- De Benedittis C, Bellan M, Crevola M, et al. Interplay of PNPLA3 and HSD17B13 Variants in Modulating the Risk of Hepatocellular Carcinoma among Hepatitis C Patients. Gastroenterol Res Pract 2020;2020:4216451. [Crossref] [PubMed]
- Bayoumi A, Elsayed A, Han S, et al. Mistranslation Drives Alterations in Protein Levels and the Effects of a Synonymous Variant at the Fibroblast Growth Factor 21 Locus. Adv Sci (Weinh) 2021;8:2004168. [Crossref] [PubMed]
- Metwally M, Bayoumi A, Romero-Gomez M, et al. A polymorphism in the Irisin-encoding gene (FNDC5) associates with hepatic steatosis by differential miRNA binding to the 3'UTR. J Hepatol 2019;70:494-500. [Crossref] [PubMed]
- Metwally M, Bayoumi A, Khan A, et al. Copy number variation and expression of exportin-4 associates with severity of fibrosis in metabolic associated fatty liver disease. EBioMedicine 2021;70:103521. [Crossref] [PubMed]
- Chang KC, Tseng PL, Wu YY, et al. A polymorphism in interferon L3 is an independent risk factor for development of hepatocellular carcinoma after treatment of hepatitis C virus infection. Clin Gastroenterol Hepatol 2015;13:1017-24. [Crossref] [PubMed]
- Salum GM, Dawood RM, Abd El-Meguid M, et al. Correlation between IL28B/TLR4 genetic variants and HCC development with/without DAAs treatment in chronic HCV patients. Genes Dis 2020;7:392-400. [Crossref] [PubMed]
- Miki D, Akita T, Kurisu A, et al. PNPLA3 and HLA-DQB1 polymorphisms are associated with hepatocellular carcinoma after hepatitis C virus eradication. J Gastroenterol 2020;55:1162-70. [Crossref] [PubMed]
- Ogawa E, Takayama K, Hiramine S, et al. Association between steatohepatitis biomarkers and hepatocellular carcinoma after hepatitis C elimination. Aliment Pharmacol Ther 2020;52:866-76. [Crossref] [PubMed]
- Thabet K, Asimakopoulos A, Shojaei M, et al. MBOAT7 rs641738 increases risk of liver inflammation and transition to fibrosis in chronic hepatitis C. Nat Commun 2016;7:12757. [Crossref] [PubMed]
- Thabet K, Chan HLY, Petta S, et al. The membrane-bound O-acyltransferase domain-containing 7 variant rs641738 increases inflammation and fibrosis in chronic hepatitis B. Hepatology 2017;65:1840-50. [Crossref] [PubMed]
- Alharthi J, Bayoumi A, Thabet K, et al. A metabolic associated fatty liver disease risk variant in MBOAT7 regulates toll like receptor induced outcomes. Nat Commun 2022;13:7430. [Crossref] [PubMed]
- Simili A, Mazzella G, Ravaioli F, et al. Interleukin 28 Polymorphisms and Hepatocellular Carcinoma Development after Direct Acting Antiviral Therapy for Chronic Hepatitis C. J Gastrointestin Liver Dis 2019;28:449-56. [Crossref] [PubMed]
- Degasperi E, Galmozzi E, Pelusi S, et al. Hepatic Fat-Genetic Risk Score Predicts Hepatocellular Carcinoma in Patients With Cirrhotic HCV Treated With DAAs. Hepatology 2020;72:1912-23. [Crossref] [PubMed]
- Eslam M, Valenti L, Romeo S. Genetics and epigenetics of NAFLD and NASH: Clinical impact. J Hepatol 2018;68:268-79. [Crossref] [PubMed]
- Mann DA. Epigenetics in liver disease. Hepatology 2014;60:1418-25. [Crossref] [PubMed]
- Feinberg AP, Koldobskiy MA, Göndör A. Epigenetic modulators, modifiers and mediators in cancer aetiology and progression. Nat Rev Genet 2016;17:284-99. [Crossref] [PubMed]
- Domovitz T, Gal-Tanamy M. Tracking Down the Epigenetic Footprint of HCV-Induced Hepatocarcinogenesis. J Clin Med 2021;10:551. [Crossref] [PubMed]
- Choi J, Corder NL, Koduru B, et al. Oxidative stress and hepatic Nox proteins in chronic hepatitis C and hepatocellular carcinoma. Free Radic Biol Med 2014;72:267-84. [Crossref] [PubMed]
- Ivanov AV, Valuev-Elliston VT, Tyurina DA, et al. Oxidative stress, a trigger of hepatitis C and B virus-induced liver carcinogenesis. Oncotarget 2017;8:3895-932. [Crossref] [PubMed]
- Wijetunga NA, Pascual M, Tozour J, et al. A pre-neoplastic epigenetic field defect in HCV-infected liver at transcription factor binding sites and polycomb targets. Oncogene 2017;36:2030-44. [Crossref] [PubMed]
- Perez S, Kaspi A, Domovitz T, et al. Hepatitis C virus leaves an epigenetic signature post cure of infection by direct-acting antivirals. PLoS Genet 2019;15:e1008181. [Crossref] [PubMed]
- Hardy T, Zeybel M, Day CP, et al. Plasma DNA methylation: a potential biomarker for stratification of liver fibrosis in non-alcoholic fatty liver disease. Gut 2017;66:1321-8. [Crossref] [PubMed]
- Shen J, Wang S, Zhang YJ, et al. Genome-wide DNA methylation profiles in hepatocellular carcinoma. Hepatology 2012;55:1799-808. [Crossref] [PubMed]
- Ammerpohl O, Pratschke J, Schafmayer C, et al. Distinct DNA methylation patterns in cirrhotic liver and hepatocellular carcinoma. Int J Cancer 2012;130:1319-28. [Crossref] [PubMed]
- Gao W, Kondo Y, Shen L, et al. Variable DNA methylation patterns associated with progression of disease in hepatocellular carcinomas. Carcinogenesis 2008;29:1901-10. [Crossref] [PubMed]
- Okamoto Y, Shinjo K, Shimizu Y, et al. Hepatitis virus infection affects DNA methylation in mice with humanized livers. Gastroenterology 2014;146:562-72. [Crossref] [PubMed]
- Matsukura S, Soejima H, Nakagawachi T, et al. CpG methylation of MGMT and hMLH1 promoter in hepatocellular carcinoma associated with hepatitis viral infection. Br J Cancer 2003;88:521-9. [Crossref] [PubMed]
- Pogribny IP, Rusyn I. Role of epigenetic aberrations in the development and progression of human hepatocellular carcinoma. Cancer Lett 2014;342:223-30. [Crossref] [PubMed]
- Song MA, Kwee SA, Tiirikainen M, et al. Comparison of genome-scale DNA methylation profiles in hepatocellular carcinoma by viral status. Epigenetics 2016;11:464-74. [Crossref] [PubMed]
- Bayoumi A, Grønbæk H, George J, et al. The Epigenetic Drug Discovery Landscape for Metabolic-associated Fatty Liver Disease. Trends Genet 2020;36:429-41. [Crossref] [PubMed]
- Hamdane N, Jühling F, Crouchet E, et al. HCV-Induced Epigenetic Changes Associated With Liver Cancer Risk Persist After Sustained Virologic Response. Gastroenterology 2019;156:2313-2329.e7. [Crossref] [PubMed]
- Zhao P, Malik S, Xing S. Epigenetic Mechanisms Involved in HCV-Induced Hepatocellular Carcinoma (HCC). Front Oncol 2021;11:677926. [Crossref] [PubMed]
- Hlady RA, Zhao X, El Khoury LY, et al. Interferon drives HCV scarring of the epigenome and creates targetable vulnerabilities following viral clearance. Hepatology 2022;75:983-96. [Crossref] [PubMed]
- Saunders MA, Liang H, Li WH. Human polymorphism at microRNAs and microRNA target sites. Proc Natl Acad Sci U S A 2007;104:3300-5. [Crossref] [PubMed]
- Liu CH, Ampuero J, Gil-Gómez A, et al. miRNAs in patients with non-alcoholic fatty liver disease: A systematic review and meta-analysis. J Hepatol 2018;69:1335-48. [Crossref] [PubMed]
- Wei XY, Ding J, Tian WG, et al. MicroRNA-122 as a diagnostic biomarker for hepatocellular carcinoma related to hepatitis C virus: a meta-analysis and systematic review. J Int Med Res 2020;48:300060520941634. [Crossref] [PubMed]
- Nomair AM, Issa NM, Madkour MA, et al. The clinical significance of serum miRNA-224 expression in hepatocellular carcinoma. Clin Exp Hepatol 2020;6:20-7. [Crossref] [PubMed]
- Rashad NM, El-Shal AS, Shalaby SM, et al. Serum miRNA-27a and miRNA-18b as potential predictive biomarkers of hepatitis C virus-associated hepatocellular carcinoma. Mol Cell Biochem 2018;447:125-36. [Crossref] [PubMed]
- Kopp F, Mendell JT. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell 2018;172:393-407. [Crossref] [PubMed]
- Sun Q, Hao Q, Prasanth KV. Nuclear Long Noncoding RNAs: Key Regulators of Gene Expression. Trends Genet 2018;34:142-57. [Crossref] [PubMed]
- Khatun M, Sur S, Steele R, et al. Inhibition of Long Noncoding RNA Linc-Pint by Hepatitis C Virus in Infected Hepatocytes Enhances Lipogenesis. Hepatology 2021;74:41-54. [Crossref] [PubMed]
- El-Khazragy N, Elshimy AA, Hassan SS, et al. lnc-HOTAIR predicts hepatocellular carcinoma in chronic hepatitis C genotype 4 following direct-acting antivirals therapy. Mol Carcinog 2020;59:1382-91. [Crossref] [PubMed]
- Refai NS, Louka ML, Halim HY, et al. Long non-coding RNAs (CASC2 and TUG1) in hepatocellular carcinoma: Clinical significance. J Gene Med 2019;21:e3112. [Crossref] [PubMed]
- Eslam M, Ampuero J, Jover M, et al. Predicting portal hypertension and variceal bleeding using non-invasive measurements of metabolic variables. Ann Hepatol 2013;12:588-98. [Crossref] [PubMed]
- Khattab MA, Eslam M, Aly MM, et al. Serum lipids and chronic hepatitis C genotype 4: interaction and significance. Ann Hepatol 2012;11:37-46. [Crossref] [PubMed]
- Khattab MA, Eslam M, Aly MM, et al. Association of serum adipocytokines with insulin resistance and liver injury in patients with chronic hepatitis C genotype 4. J Clin Gastroenterol 2012;46:871-9. [Crossref] [PubMed]
- Khattab MA, Eslam M, Shatat M, et al. Changes in adipocytokines and insulin sensitivity during and after antiviral therapy for hepatitis C genotype 4. J Gastrointestin Liver Dis 2012;21:59-65. [PubMed]
- Khattab MA, Eslam M, Mousa YI, et al. Association between metabolic abnormalities and hepatitis C-related hepatocellular carcinoma. Ann Hepatol 2012;11:487-94. [Crossref] [PubMed]
- Fouad Y, Lazarus JV, Negro F, et al. MAFLD considerations as a part of the global hepatitis C elimination effort: an international perspective. Aliment Pharmacol Ther 2021;53:1080-9. [Crossref] [PubMed]
- Chen S, Zhang Y, Wu X, et al. Diagnostic Value of lncRNAs as Biomarker in Hepatocellular Carcinoma: An Updated Meta-Analysis. Can J Gastroenterol Hepatol 2018;2018:8410195. [Crossref] [PubMed]
- Lin XJ, Chong Y, Guo ZW, et al. A serum microRNA classifier for early detection of hepatocellular carcinoma: a multicentre, retrospective, longitudinal biomarker identification study with a nested case-control study. Lancet Oncol 2015;16:804-15. [Crossref] [PubMed]
- Yang J, Sheng YY, Wei JW, et al. MicroRNA-219-5p Promotes Tumor Growth and Metastasis of Hepatocellular Carcinoma by Regulating Cadherin 1. Biomed Res Int 2018;2018:4793971. [Crossref] [PubMed]
- Cho Y, Kim BH, Park JW. Overview of Asian clinical practice guidelines for the management of hepatocellular carcinoma: An Asian perspective comparison. Clin Mol Hepatol 2023;29:252-62. [Crossref] [PubMed]
- Eslam M, George J. Genetic Insights for Drug Development in NAFLD. Trends Pharmacol Sci 2019;40:506-16. [Crossref] [PubMed]
- Cook D, Brown D, Alexander R, et al. Lessons learned from the fate of AstraZeneca's drug pipeline: a five-dimensional framework. Nat Rev Drug Discov 2014;13:419-31. [Crossref] [PubMed]
- Nelson MR, Tipney H, Painter JL, et al. The support of human genetic evidence for approved drug indications. Nat Genet 2015;47:856-60. [Crossref] [PubMed]
- Zhang B, Jia WH, Matsuda K, et al. Large-scale genetic study in East Asians identifies six new loci associated with colorectal cancer risk. Nat Genet 2014;46:533-42. [Crossref] [PubMed]
- Al Olama AA, Kote-Jarai Z, Berndt SI, et al. A meta-analysis of 87,040 individuals identifies 23 new susceptibility loci for prostate cancer. Nat Genet 2014;46:1103-9. [Crossref] [PubMed]
- Bishop DT, Demenais F, Iles MM, et al. Genome-wide association study identifies three loci associated with melanoma risk. Nat Genet 2009;41:920-5. [Crossref] [PubMed]
- Asgari MM, Wang W, Ioannidis NM, et al. Identification of Susceptibility Loci for Cutaneous Squamous Cell Carcinoma. J Invest Dermatol 2016;136:930-7. [Crossref] [PubMed]
- Chahal HS, Wu W, Ransohoff KJ, et al. Genome-wide association study identifies 14 novel risk alleles associated with basal cell carcinoma. Nat Commun 2016;7:12510. [Crossref] [PubMed]
- Kelly TK, De Carvalho DD, Jones PA. Epigenetic modifications as therapeutic targets. Nat Biotechnol 2010;28:1069-78. [Crossref] [PubMed]
- Bitzer M, Horger M, Giannini EG, et al. Resminostat plus sorafenib as second-line therapy of advanced hepatocellular carcinoma - The SHELTER study. J Hepatol 2016;65:280-8. [Crossref] [PubMed]
- He B, Dai L, Zhang X, et al. The HDAC Inhibitor Quisinostat (JNJ-26481585) Supresses Hepatocellular Carcinoma alone and Synergistically in Combination with Sorafenib by G0/G1 phase arrest and Apoptosis induction. Int J Biol Sci 2018;14:1845-58. [Crossref] [PubMed]
- Bárcena-Varela M, Caruso S, Llerena S, et al. Dual Targeting of Histone Methyltransferase G9a and DNA-Methyltransferase 1 for the Treatment of Experimental Hepatocellular Carcinoma. Hepatology 2019;69:587-603. [Crossref] [PubMed]
- Jühling F, Hamdane N, Crouchet E, et al. Targeting clinical epigenetic reprogramming for chemoprevention of metabolic and viral hepatocellular carcinoma. Gut 2021;70:157-69. [Crossref] [PubMed]


